Title
题目
Universal and extensible language-vision models for organ segmentation and tumor detection from abdominal computed tomography
用于腹部计算机断层扫描中器官分割和肿瘤检测的通用且可扩展的语言-视觉模型
01
文献速递介绍
计算机断层扫描(CT)是一种广泛使用且功能强大的疾病诊断和治疗计划工具(Mattikalli 等,2022;Zhou 等,2022;Qu 等,2023;Chen 等,2024a)。在常规的临床工作流程中,放射科医生需要分析单个CT卷中的数百个2D切片,以寻找和解读诊断信息,这个过程既繁琐又容易导致误诊(Zhou,2021)。医学图像分割提供了一个有前景的解决方案,通过自动识别器官、描绘其边界并突出显示异常,来提高诊断效率和质量(Liu 等,2021;Hu 等,2023;Chen 等,2023a,2024b;Lai 等,2024)。
医学图像分割的进展在很大程度上依赖于专门的数据集。这些数据集包括器官/肿瘤特定的数据集,例如LiTS(肝肿瘤分割)(Bilic 等,2019)、KiTS(肾肿瘤分割)(Heller 等,2019)和MSD(医学分割十项全能)(Simpson 等,2019),以及腹部多器官标注数据集,如BTCV(Beyond The Cranial Vault)(Landman 等,2015)、AMOS(腹部多器官分割)(Ji 等,2022)和AbdomenAtlas(Li 等,2024)。此外,Wasserthal 等(2023)提出了一种提供全身解剖视图的方法,旨在捕捉人体的综合解剖结构,而不仅仅是特定的身体区域。
考虑到人体解剖结构的高度复杂性和对精细临床要求的需求,我们可以预见新的器官/肿瘤标注的出现(Jaus 等,2023),例如阑尾和脾脏肿瘤的标注。此外,对更详细的解剖标注的需求也在增长,例如区分肝脏的右叶和左叶(Germain 等,2014)。虽然在当前的数据集中,肝脏的标注很常见,但肝叶的划分研究仍然不足(Bilic 等,2023),这可能导致未来肝叶与整个肝脏之间标注的重叠。为满足这些新兴需求,最新的努力涉及使用人类参与重新标注现有数据集并相应地重新训练模型(Qu 等,2023;Wasserthal 等,2023;Jaus 等,2023)。然而,这种方法带来了巨大的标注成本,特别是在3D医学影像中,并且重新从头训练模型还需要大量的计算资源(Zhang 等,2023b,2024)。因此,探索一种能够有效处理新器官/肿瘤标注并减轻重新训练模型所带来的计算负担的新框架显得尤为重要。
Abatract
摘要
The advancement of artificial intelligence (AI) for organ segmentation and tumor detection is propelled by thegrowing availability of computed tomography (CT) datasets with detailed, per-voxel annotations. However,these AI models often struggle with flexibility for partially annotated datasets and extensibility for new classesdue to limitations in the one-hot encoding, architectural design, and learning scheme. To overcome theselimitations, we propose a universal, extensible framework enabling a single model, termed Universal Model,to deal with multiple public datasets and adapt to new classes (e.g., organs/tumors). Firstly, we introduce anovel language-driven parameter generator that leverages language embeddings from large language models,enriching semantic encoding compared with one-hot encoding. Secondly, the conventional output layers arereplaced with lightweight, class-specific heads, allowing Universal Model to simultaneously segment 25 organsand six types of tumors and ease the addition of new classes. We train our Universal Model on 3410 CTvolumes assembled from 14 publicly available datasets and then test it on 6173 CT volumes from four externaldatasets. Universal Model achieves first place on six CT tasks in the Medical Segmentation Decathlon (MSD)public leaderboard and leading performance on the Beyond The Cranial Vault (BTCV) dataset. In summary,Universal Model exhibits remarkable computational efficiency (6× faster than other dataset-specific models),demonstrates strong generalization across different hospitals, transfers well to numerous downstream tasks,and more importantly, facilitates the extensibility to new classes while alleviating the catastrophic forgetting of previously learned classes. Codes,
人工智能(AI)在器官分割和肿瘤检测方面的进步得益于带有详细体素级注释的计算机断层扫描(CT)数据集的日益普及。然而,由于单热编码、架构设计和学习方案的限制,这些AI模型通常在处理部分标注数据集以及扩展到新类别(例如器官/肿瘤)时表现出灵活性不足。为克服这些限制,我们提出了一个通用且可扩展的框架,使单一模型(称为Universal Model)能够处理多个公共数据集并适应新类别。首先,我们引入了一种新颖的语言驱动参数生成器,该生成器利用大语言模型中的语言嵌入,与单热编码相比,增强了语义编码。其次,我们将传统的输出层替换为轻量级的类别特定头部,使Universal Model能够同时分割25个器官和6种类型的肿瘤,并简化新类别的添加。我们在由14个公开可用数据集组成的3410个CT卷上训练了Universal Model,并在来自四个外部数据集的6173个CT卷上对其进行了测试。Universal Model在医学分割十项全能(MSD)公开排行榜的六项CT任务中获得第一名,并在Beyond The Cranial Vault(BTCV)数据集上取得领先表现。总而言之,Universal Model展现了卓越的计算效率(比其他特定数据集的模型快6倍)、在不同医院之间的强泛化能力、良好的下游任务迁移性,更重要的是,它在扩展新类别时有效避免了先前学习类别的灾难性遗忘。
Method
方法
Let us consider a set of partially labeled datasets {1 , 2 , …, 𝑁 }with corresponding organs/tumors label space {1 ,2 , …,𝑁 }, welearn a unified multi-organ segmentation model to segment tumors/organs in = ⋃𝑁𝑖𝑖 , where ‖‖ ≥ ‖𝑖‖ for all 𝑖 ∈ {1, 2, … , 𝑁}. Whenit comes a new dataset 𝑁+1 with organs/tumors label space 𝑁+1,𝑁+1 ⧵ ≠ ∅, all previous training data are not accessible. The modelis required to predict the accumulated label space ⋃ 𝑁+1 for all seendatasets {1 , 2 , …, 𝑁 , 𝑁+1}.
假设我们有一组部分标注的数据集{1 , 2 , …, 𝑁* },以及与其对应的组织/肿瘤标签空间{1 ,2 , …,𝑁 },我们学习一个统一的多组织分割模型,用于在 = ⋃𝑁**𝑖𝑖中分割肿瘤/组织,其中对于所有𝑖∈{1, 2, … , 𝑁},都有‖‖≥‖𝑖‖。当遇到一个新的数据集𝑁+1,其组织/肿瘤标签空间为𝑁+1,且𝑁+1⧵⧵≠∅,此时所有之前的训练数据不可用。模型需要预测所有已见过的数据集{1 , 2 , …, 𝑁 , 𝑁+1}的累计标签空间⋃𝑁+1。
Conclusion
结论
In this work, we present a novel framework for multi-organ segmentation and tumor detection. This work integrates language embedding with segmentation models to enable a flexible and powerfulsegmentor, allowing the model to learn from an integrated datasetfor high-performing organ segmentation and tumor detection, ranking first in both MSD and BTCV. More importantly, we demonstratethat language embedding can establish the anatomical relationship. Finally, the experiment results validate several clinically important meritsof the CLIP-Driven Universal Model, such as compelling efficiency,generalizability, transferability, and extensibility.
在这项工作中,我们提出了一个新颖的框架,用于多脏器分割和肿瘤检测。该框架将语言嵌入与分割模型集成,以实现灵活且强大的分割器,使模型能够从综合数据集中学习,从而在器官分割和肿瘤检测方面表现出色,在 MSD 和 BTCV 中均排名第一。更重要的是,我们展示了语言嵌入如何建立解剖关系。最后,实验结果验证了 CLIP 驱动的通用模型在效率、泛化能力、迁移能力和扩展性等临床重要特性方面的显著优势。
Figure
图
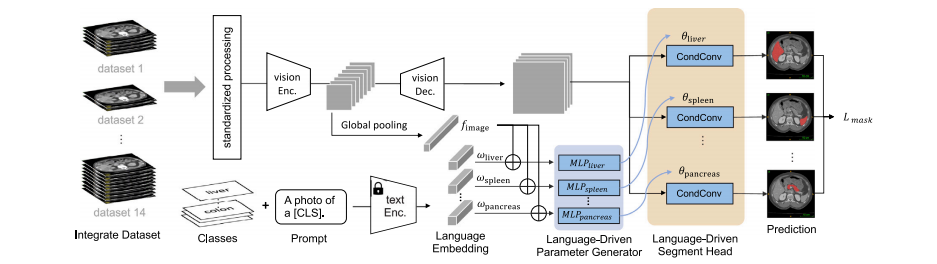
Fig. 1. Overview. We have developed the continual CLIP-Driven Universal Model from an assembly of 14 public datasets of 3410 CT volumes. In total, 25 organs and 6 types oftumors are partially labeled. To deal with partial labels, Universal Model consists of a language branch and a vision branch (Section 3.2). The official test set of MSD and BTCVare used to benchmark the performance of organ segmentation (Section 4.2) and tumor detection (Section 4.3). 3D-IRCADb, TotalSegmentator and a large-scale private dataset,consisting of 5038 CT volumes with 21 annotated organs, are used for independent, external validation of model generalizability (Section 4.5) and transferability (Section 4.6).The LPG module utilizes separate MLPs for each organ to overcome the entanglement issue present in the ICCV version (Liu et al., 2023a), which relied on a single MLP.
图1. 概述. 我们从14个公共数据集的3410个CT卷中开发了持续CLIP驱动的通用模型。总共有25个器官和6种类型的肿瘤部分标注。为了处理部分标注,通用模型包括语言分支和视觉分支(第3.2节)。MSD和BTCV的官方测试集用于器官分割(第4.2节)和肿瘤检测(第4.3节)的性能基准测试。3D-IRCADb、TotalSegmentator和一个包含5038个CT卷的大规模私人数据集(包括21个标注器官)用于模型泛化能力(第4.5节)和可迁移性(第4.6节)的独立外部验证。LPG模块利用每个器官的独立MLP来克服ICCV版本中存在的纠缠问题(Liu 等,2023a),该版本依赖于单一的MLP。
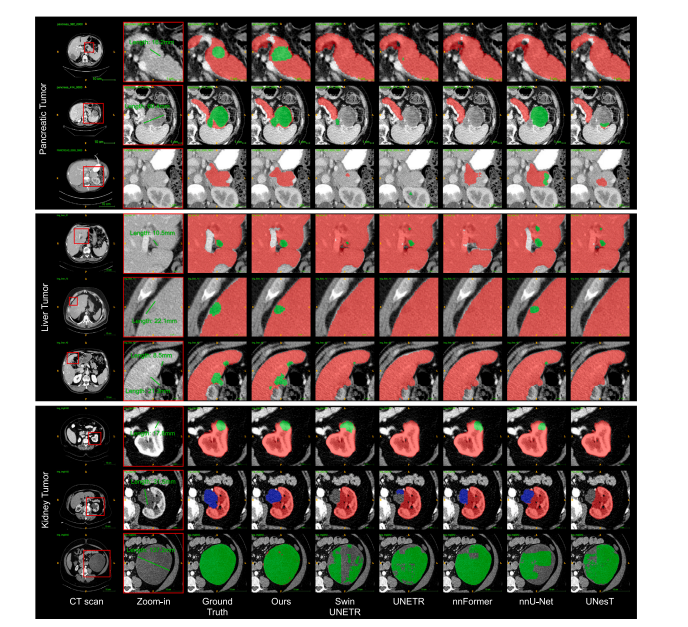
Fig. 2. Qualitative results of multi-tumor detection and segmentation. We review the detection/segmentation results of each tumor type from smaller to larger sizes. Especially,Universal Model generalizes well in organ segmentation and does not generate many false positives of tumors when it comes to a CT volume without tumors from other hospitals(Row 3).
图2. 多肿瘤检测和分割的定性结果。我们按从小到大的顺序回顾了每种类型肿瘤的检测/分割结果。特别是,通用模型在器官分割中表现出良好的泛化能力,并且在处理来自其他医院的无肿瘤CT卷时不会产生许多肿瘤的假阳性(第3行)。
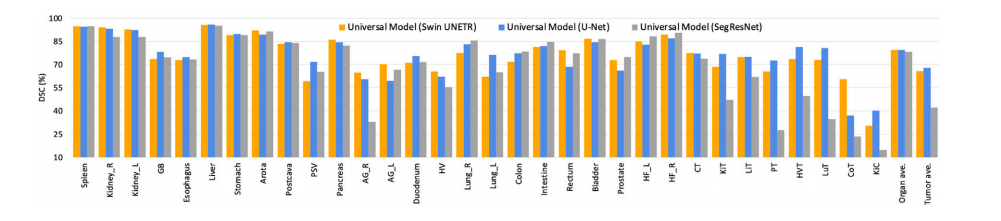
Fig. 3. Ablation study on different segmentation backbones. Universal Model can be expanded to Transformer-based (Swin UNETR) and CNN-based (U-Net, SegResNet-Tiny) backbone.These backbones achieve comparable results. The numbers of parameters of Swin UNETR, U-Net, and SegResNet-Tiny are 62.19M, 19.08M, and 4.7M, respectively. The order ofclasses is the same as Table 2.
图3. 不同分割主干网络的消融研究。通用模型可以扩展到基于Transformer的(Swin UNETR)和基于CNN的(U-Net, SegResNet-Tiny)主干网络。这些主干网络取得了相当的结果。Swin UNETR、U-Net和SegResNet-Tiny的参数数量分别为62.19M、19.08M和4.7M。类别的顺序与表2相同。
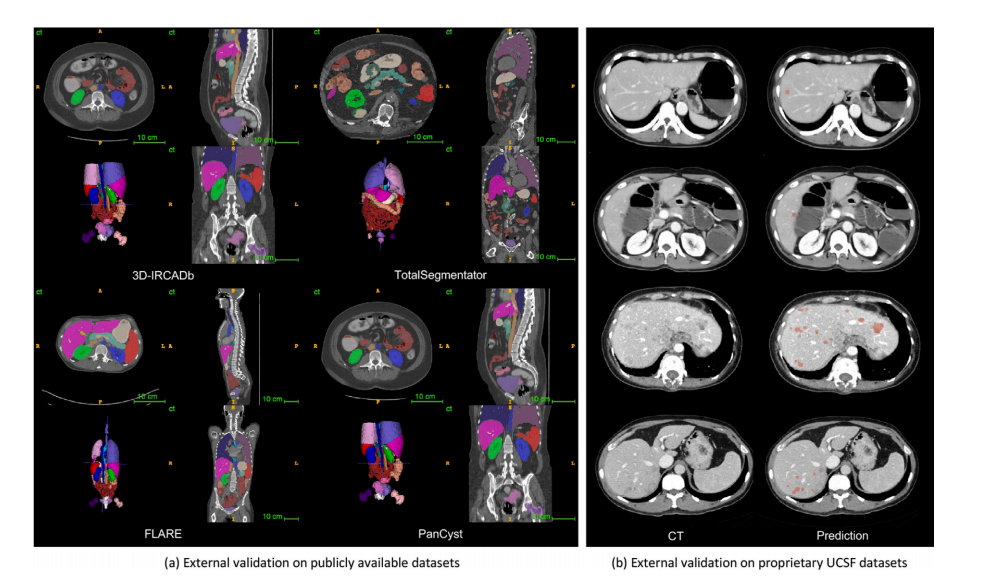
Fig. 4. (a) Pseudo-label visualization. 25 organs and 6 tumors in four unseen datasets are visualized. (b) External validation for liver tumor detection. In Cases 1 and 2, UniversalModel successfully identified small new liver tumors, that have been overlooked during radiological evaluation. In Cases 3 and 4, where multiple liver tumors were present,Universal Model detected them, resulting in improved diagnostic efficiency.
图4. (a) 伪标签可视化。在四个未见过的数据集中可视化了25个器官和6个肿瘤。(b) 肝肿瘤检测的外部验证。在病例1和2中,通用模型成功识别出放射评估中被忽略的小型新肝肿瘤。在病例3和4中,通用模型检测到了多个肝肿瘤,提高了诊断效率。

Fig. 5. (a) Contour line comparison among pseudo labels and two human experts. The red line represents the annotation from Doctor 1; green line indicates the annotation fromDoctor 2; blue line shows the results generated by Universal Model. Examples of CT volumes annotated by our pseudo labels and two human experts with contour line comparison.The prediction results of these organs generated by the medical model are comparable with human experts. (b) Intra-observer variability. We obtain similar performance betweenpseudo labels generated by Universal Model (AI) and annotations performed by two human experts (Dr1,2) on 6 organs. Spleen (Spl), liver (Liv), kidneys (Kid), stomach (Sto),gallbladder (Gall), and pancreas (Pan) can be annotated by AI with a similar intra-observer variability to humans.
图5. (a) 伪标签与两位专家之间的轮廓线比较。红线代表医生1的标注;绿线表示医生2的标注;蓝线显示由通用模型生成的结果。展示了使用我们的伪标签和两位专家标注的CT卷样例以及轮廓线比较。医学模型生成的这些器官的预测结果与人类专家相当。(b) 观察者内变异性。我们在6个器官上获得了通用模型(AI)生成的伪标签与两位专家(Dr1,2)标注之间的相似表现。脾脏(Spl)、肝脏(Liv)、肾脏(Kid)、胃(Sto)、胆囊(Gall)和胰腺(Pan)的标注结果显示,AI的观察者内变异性与人类相似。
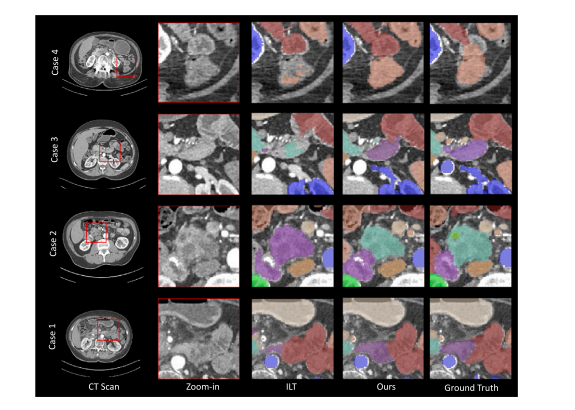
Fig. 6. The visualization for body extension benchmark. We compare Continual Universal Model with baseline model ILT across four case studies. Our model demonstrates effectiveadaptation to new organs while retaining knowledge of previously learned classes, thereby avoiding forgetting.
图6. 身体扩展基准的可视化。我们在四个案例研究中比较了持续学习的通用模型与基线模型ILT。我们的模型在适应新器官的同时保留了先前学习类别的知识,从而避免了遗忘
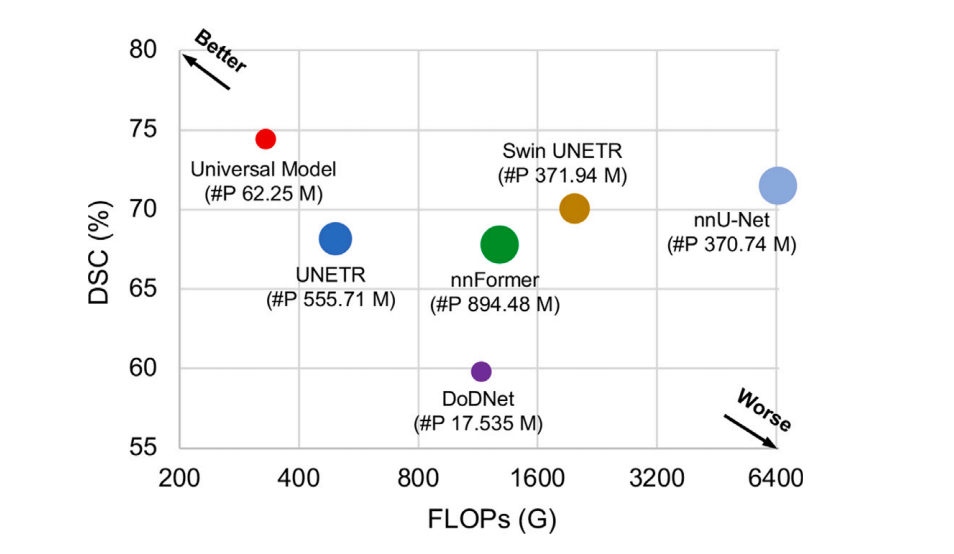
Fig. 7. Inference speed with FLOPs vs. DSC. The plot illustrates the relationship between the average DSC similarity coefficient (DSC) score across the six Medical SegmentationDecathlon (MSD) tasks and the floating-point operations per second (FLOPs). The FLOPs are calculated based on input with a spatial size of 96 × 96 × 96. The size of each circlein the plot corresponds to the number of parameters (‘#P’). The used backbone of Universal Model is Swin UNETR.
图7. 推理速度与FLOPs对比DSC。该图展示了六项医学分割十项全能(MSD)任务中平均DSC相似系数(DSC)得分与每秒浮点运算次数(FLOPs)之间的关系。FLOPs是基于空间尺寸为96 × 96 × 96的输入计算的。图中每个圆的大小对应于参数的数量(‘#P’)。通用模型使用的主干网络是Swin UNETR。
Table
表

Table 1The information for an assembly of datasets.* We have developed Universal Model from an assembly of 1–14 public datasets. The official test and validation sets of Medical SegmentationDecathlon (MSD) and Beyond the Cranial Vault (BTCV) are used to benchmark the performance of organ segmentation (Section 4.2) and tumor detection (Section 4.3). 3D-IRCADb(15), TotalSegmentator (16) and a large-scale private dataset (17), consisting of 5038 CT volumes with 21 annotated organs, are used for independent evaluation of modelgeneralizability (Section 4.5) and transferability (Section 4.6).
表1数据集集合的信息。我们从1到14个公共数据集的集合中开发了通用模型。医学分割十项全能(MSD)和Beyond the Cranial Vault(BTCV)的官方测试和验证集用于器官分割(第4.2节)和肿瘤检测(第4.3节)的性能基准测试。3D-IRCADb(15)、TotalSegmentator(16)和一个包含5038个CT卷的大规模私人数据集(17),其中包含21个标注器官,均用于模型泛化能力(第4.5节)和可迁移性(第4.6节)的独立评估。
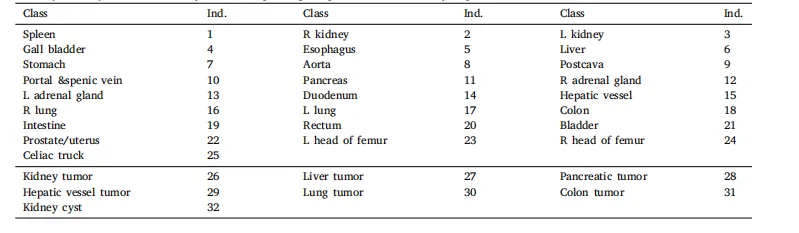
Table 2The unification of the label taxonomy. The corresponding template for this taxonomy is presented, where ind. denotes the index of the class.
表2 标签分类的统一化。本表展示了该分类法的对应模板,其中ind.表示类别的索引。

Table 3Leaderboard performance of CT tasks on MSD. The results are evaluated in the server on the MSD competition test dataset. The dice similarity coefficient (DSC) and normalizedsurface distance (NSD) metrics are obtained from the MSD public leaderboard. The results of MRI-related tasks were generated by nnU-Net (Isensee et al., 2021).
表3 MSD上CT任务的排行榜表现。结果在MSD竞赛测试数据集的服务器上进行评估。Dice相似系数(DSC)和归一化表面距离(NSD)指标是从MSD公共排行榜上获得的。MRI相关任务的结果由nnU-Net(Isensee 等,2021)生成。
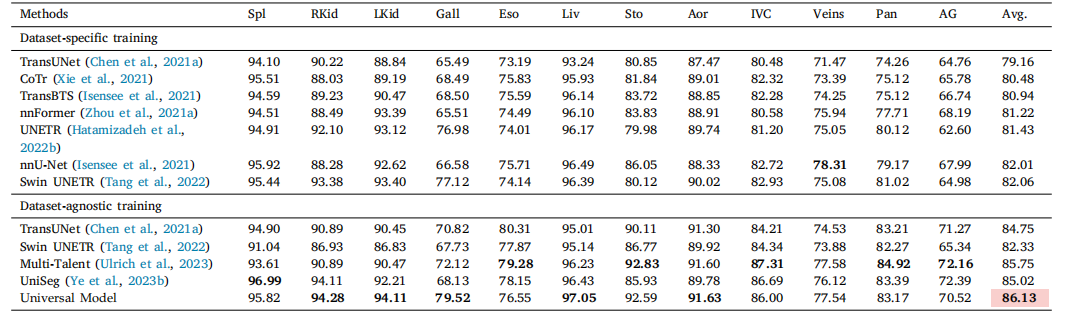
Table 4Five-fold cross-validation results on BTCV.* For a fair comparison, we did not use model ensemble during the evaluation. The dataset-specific training means train only on BTCV,while dataset-agnostic training means training on multiple datasets. In dataset-agnostic training part, results of Swin UNETR and Multi-Talent are original from Ulrich et al. (2023).We re-implement TansUNet and UniSeg with same setting as Universal Model. Universal Model achieves the overall best performance. The performance is statistically significantat the P = 0.05 level, with highlighting in a light red box.
表4BTCV上的五折交叉验证结果。为了公平比较,在评估过程中我们没有使用模型集成。数据集特定训练是指仅在BTCV上训练,而数据集无关训练是指在多个数据集上进行训练。在数据集无关训练部分,Swin UNETR和Multi-Talent的结果来自Ulrich等(2023)。我们在相同设置下重新实现了TansUNet和UniSeg与通用模型的比较。通用模型取得了整体最佳表现。性能在P = 0.05水平上具有统计显著性,结果在浅红色框中突出显示。

Table 5Benchmark of different partially labeled methods on MOTS dataset. Universal Model* denotes the model trained exclusively on the MOTS training set, while Universal Model refersto our proposed model trained on the 14 integrated datasets in this work, excluding the 235 CT volumes present in the MOTS test set. The average performance is statisticallysignificant at the P = 0.05 level, with highlighting in a light red box.
表5 不同部分标注方法在MOTS数据集上的基准测试。Universal Model 指的是仅在MOTS训练集上训练的模型,而Universal Model指的是本文中提出的在14个集成数据集上训练的模型,不包括MOTS测试集中存在的235个CT卷。平均性能在P = 0.05水平上具有统计显著性,结果在浅红色框中突出显示。

Table 6Quantitative results of multi-tumor detection. The tumor detection performance analysis is based on CT volumes from the LiTS, KiTS, and MSD Pancreas datasets, containing tumors inthe liver, kidney, and pancreas, respectively (Bilic et al., 2019; Heller et al., 2019; Antonelli et al., 2021). These volumes are utilized to compute the sensitivity of tumor detection.To assess specificity in an alternative manner, the CHAOS and Pancreas-CT datasets are employed (Valindria et al., 2018; Roth et al., 2015). Importantly, it has been confirmedthat the CHAOS dataset is devoid of liver or kidney tumors, while the Pancreas-CT dataset does not contain pancreatic tumors within the CT volumes. Universal Model achievesa high harmonic mean, which signifies the capability to accurately identify tumor cases while minimizing false positives.
表6 多肿瘤检测的定量结果。肿瘤检测性能分析基于LiTS、KiTS和MSD胰腺数据集的CT卷,这些数据集分别包含肝脏、肾脏和胰腺中的肿瘤(Bilic 等,2019;Heller 等,2019;Antonelli 等,2021)。这些CT卷被用于计算肿瘤检测的敏感性。为了以另一种方式评估特异性,使用了CHAOS和Pancreas-CT数据集(Valindria 等,2018;Roth 等,2015)。重要的是,已确认CHAOS数据集没有肝脏或肾脏肿瘤,而Pancreas-CT数据集中不包含胰腺肿瘤。通用模型实现了较高的调和平均值,这表明它能够准确识别肿瘤病例,同时将假阳性降到最低。
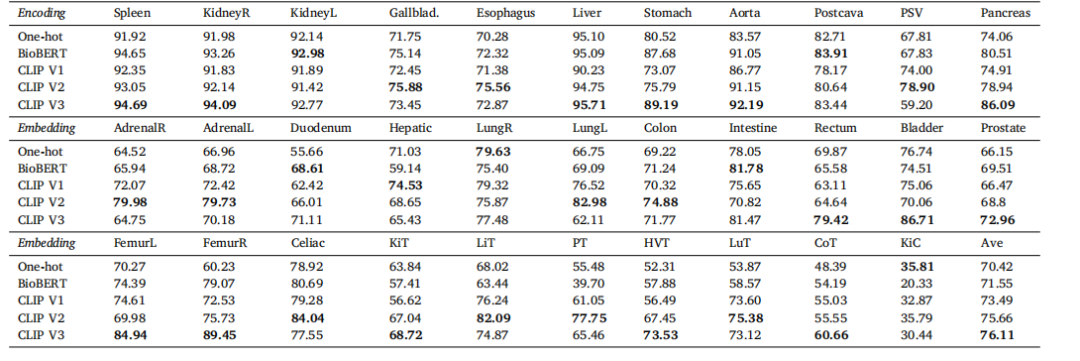
Table 7Ablation study on language encoding.* We compare the language embedding among one-hot, BioBERT (Yasunaga et al., 2022) and CLIP embedding in the test set of integrated data.The BioBERT method utilizes the prompt ‘A computerized tomography of a [CLS]’. On the other hand, CLIP v1, v2, and v3 employ prompts such as ‘A photo of a [CLS]’, ‘There is[CLS] in this computerized tomography’*, and ‘A computerized tomography of a [CLS]’, respectively.
表7 语言编码的消融研究。我们在集成数据的测试集中比较了一热编码、BioBERT(Yasunaga 等,2022)和CLIP嵌入的语言嵌入。BioBERT方法使用了提示语‘A computerized tomography of a [CLS]’。另一方面,CLIP v1、v2和v3分别使用了提示语‘A photo of a [CLS]’、‘There is [CLS] in this computerized tomography’和‘A computerized tomography of a [CLS]’。
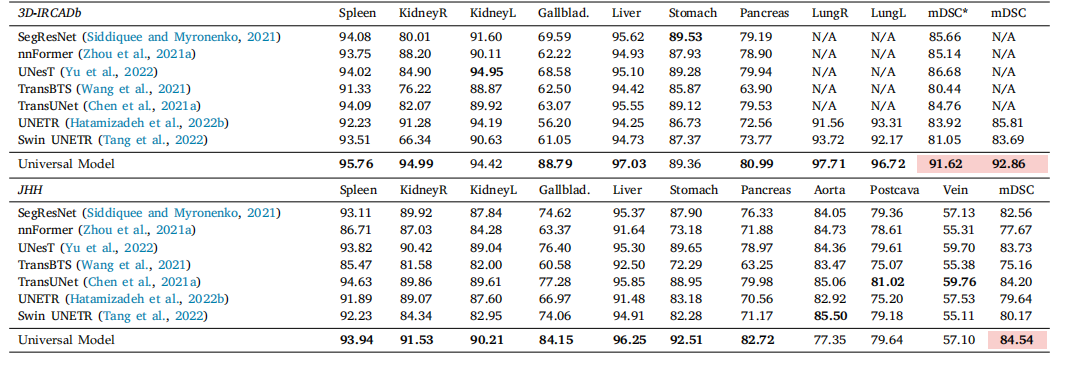
Table 8Generalizability. Results on external datasets. We evaluate Universal Model and eight models on data from two external sources without additional fine-tuning or domain adaptation.mDSC is the average DSC score of the first seven organs. Compared with dataset-specific models, our Universal Model performs more robustly to CT volumes taken from a varietyof scanners, protocols, and institutes. The average performance is statistically significant at the P = 0.05 level, with highlighting in a light red box.
表8 泛化能力。外部数据集的结果。我们在两个外部数据源的数据上评估了通用模型和八个模型,未进行额外的微调或领域适应。mDSC是前七个器官的平均DSC评分。与数据集特定的模型相比,我们的通用模型在来自各种扫描仪、协议和机构的CT卷上表现得更为稳健。平均性能在P = 0.05水平上具有统计显著性,结果在浅红色框中突出显示。

Table 9Transferability: Fine-tuning performance. Comparing with MedicalNet (Chen et al., 2019b), Models Gen. (Zhou et al., 2019c), Swin UNETR (Tang et al., 2022), UniMiSS (Xie et al.,2022b), fine-tuning Universal Model significantly outperforms learning from scratch on several downstream tasks (i.e., , vertebrae, cardiac, muscles, organs and sub-classes ofpancreatic tumor segmentation). Moreover, Universal Model, trained by image segmentation as proxy task, can extract better visual representation—more related to segmentationtasks—than other pre-trained models developed in the medical domain. The average performance is statistically significant at the P = 0.05 level, with highlighting in a light redbox.
表9 可迁移性:微调性能。与MedicalNet(Chen 等,2019b)、Models Gen.(Zhou 等,2019c)、Swin UNETR(Tang 等,2022)、UniMiSS(Xie 等,2022b)相比,微调通用模型在多个下游任务(如椎骨、心脏、肌肉、器官及胰腺肿瘤亚类分割)上显著优于从头学习。此外,通过图像分割作为代理任务训练的通用模型,能够比其他在医学领域开发的预训练模型提取出与分割任务更相关的视觉表示。平均性能在P = 0.05水平上具有统计显著性,结果在浅红色框中突出显示。
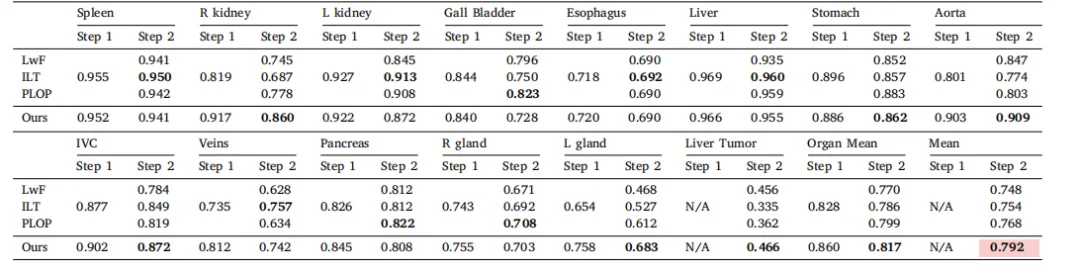
Table 10Results in tumor extension benchmark.* We present the DSC score of each class in two continual learning steps. The Organ Mean represents the average DSC score for 13 organs,and Mean stands for the average score for 14 classes. The average performance is statistically significant at the P = 0.05 level, with highlighting in a light red box.
表10 肿瘤扩展基准测试结果。我们展示了在两个持续学习步骤中每个类别的DSC评分。Organ Mean代表13个器官的平均DSC评分,Mean代表14个类别的平均评分。平均性能在P = 0.05水平上具有统计显著性,结果在浅红色框中突出显示。

Table 11Results in body extension benchmark and ablation study. We present the average DSCscore for three body parts in each step separately. The Ours_1-hot replace the languageembedding with one-hot embedding.
表 11 身体扩展基准测试和消融研究结果。我们分别展示了每一步中三个身体部位的平均 DSC 分数。Ours_1-hot 方法用 one-hot 嵌入替代了语言嵌入。