01
文献速递介绍
Medical image segmentation is a key step in medical research, diagnosis, treatment, and surgical planning. A single 3D medical image, such as a CT or an MRI scan, can be up to hundreds of megabytes in size [1]. Two-dimensional images such as radiographs or digital specimen slides are often thousands of pixels in width and height. These large sizes of images present a challenge for deep learning methods. Training on larger images requires a larger amount of GPU memory and higher-capacity neural networks [2], limiting the maximum batch size and resulting in slower convergence and a worse gradient estimate. Commonly, medical images are downscaled as a pre-processing step for medical image segmentation. This leads to a loss of fine details that are often important for accurate segmentation and consequently to a reduced segmentation accuracy [3]. In this paper, we present a new approach to training segmentation convolutional neural networks (CNNs) on very small input sizes. Our approach, which we call Segment-thensegment, is based on cropping instead of downscaling. Thus, it maintains the information content in salient regions of the image. This method does not require changing the network architecture or capacity. The method we present can be used as a general preprocessing step for any kind of segmentation neural network. Our approach uses two neural networks with small input sizes. The first network performs a rough segmentation on a uniformly downsampled input image. This rough segmentation is used to obtain salient crops of the original high-resolution image.
医学图像分割是医学研究、诊断、治疗和手术规划中的关键步骤。单个3D医学图像,如CT或MRI扫描,大小可达数百兆字节。二维图像,如X光片或数字标本幻灯片,宽度和高度通常有数千像素。这些大尺寸的图像对深度学习方法提出了挑战。在较大的图像上训练需要更多的GPU内存和更高容量的神经网络,这限制了最大批量大小,并导致收敛速度更慢和梯度估计更差。通常,医学图像在进行医学图像分割之前会被降采样作为预处理步骤。这导致了细节的丢失,而这些细节通常对于准确的分割非常重要,从而导致分割精度降低。在本文中,我们提出了一种在非常小的输入尺寸上训练分割卷积神经网络(CNNs)的新方法。我们称之为“先切割再分割”的方法,基于裁剪而不是降采样。因此,它保留了图像显著区域的信息内容。这种方法不需要改变网络架构或容量。我们提出的方法可以作为任何类型分割神经网络的通用预处理步骤。我们的方法使用两个小输入尺寸的神经网络。第一个网络在均匀降采样的输入图像上进行粗略分割。这个粗略分割用于获取原始高分辨率图像的显著裁剪区域。
Title
题目
Segment-then-Segment:Context-Preserving Crop-Based Segmentation for Large Biomedical Images
“先切割再分割:基于裁剪的大型生物医学图像分割中的上下文保留”
Abstract
摘要
Medical images are often of huge size, which presents a challenge in terms of memory requirements when training machine learning models. Commonly, the images are downsampled to overcome this challenge, but this leads to a loss of information. We present a general approach for training semantic segmentation neural networks on much smaller input sizes called Segmentthen-Segment. To reduce the input size, we use image crops instead of downscaling. One neural network performs the initial segmentation on a downscaled image. This segmentation is then used to take the most salient crops of the full-resolution image with the surrounding context. Each crop is segmented using a second specially trained neural network. The segmentation masks of each crop are joined to form the final output image. We evaluate our approach on multiple medical image modalities (microscopy, colonoscopy, and CT) and show that this approach greatly improves segmentation performance with small network input sizes when compared to baseline models trained on downscaled images, especially in terms of pixel-wise recall.
医学图像通常具有巨大的尺寸,这在训练机器学习模型时对内存要求方面提出了挑战。通常,为了克服这一挑战,图像会被下采样,但这导致了信息的丢失。我们提出了一种称为“先切割再分割”的通用方法,用于在更小的输入尺寸上训练语义分割神经网络。为了减少输入尺寸,我们使用图像裁剪而不是降低分辨率。一个神经网络在降低分辨率的图像上执行初始分割。然后,使用此分割来取得全分辨率图像中最显著的裁剪区域及其周围上下文。每个裁剪区域使用第二个特别训练的神经网络进行分割。每个裁剪区域的分割掩膜被合并形成最终输出图像。我们在多种医学图像模式(显微镜、结肠镜和CT)上评估了我们的方法,并展示了与在降低分辨率的图像上训练的基线模型相比,当输入尺寸较小时,这种方法在提高分割性能方面,特别是在像素级召回率方面,有很大的改进。
Methods
方法
A visual summary of our approach is shown in Figure 1. In segmentation CNNs, commonly the image is uniformly downsampled and then segmented. This reduces the amount of information available to the network since salient and non-salient pixels are equally downsampled. Instead of uniformly downsampling the image, we crop each object in the original high-resolution image and segment the cropped images separately. The crop regions are usually much smaller than the whole image, leading to a reduction in the network input size without losing pixel information inside the objects.
图1展示了我们方法的视觉总结。在分割CNN中,通常图像会被均匀地下采样然后进行分割。这减少了网络可用的信息量,因为显著和非显著像素都被等量地下采样。我们的方法不是均匀地下采样图像,而是在原始高分辨率图像中裁剪出每个对象,并单独对这些裁剪后的图像进行分割。裁剪区域通常比整个图像小得多,这样可以减少网络输入的大小,同时不丢失对象内的像素信息。
Results
结果
We evaluate our approach on three separate datasets described in detail in Section 3.1, hereafter referred to as the cells, aorta, and polyp datasets. First, for each dataset, we trained a rough segmentation U-Net, Res-U-Net++, and DeepLabv3+ network at various downscaled input resolutions. These models are also used as baseline models to compare against our approach. To evaluate our approach, we train fine segmentation models using the same combinations of datasets, architectures, and input sizes. Altogether more than 100 neural networks were trained to evaluate our approach, including both the baseline networks and networks trained on cropped images. Each network is trained from scratch using the downscaled dataset. The outputs from the networks are then upscaled to the datasets’ original resolution, and the metrics are calculated using those outputs. We use the held-out test datasets for all of the results reported in this section. The baseline models are used as the rough segmentation networks for the experiments using our approach. The hyperparameters used for each network are reported in Table 1. In the interest of providing objective metrics of model performance, all of the hyperparameters were tuned using the validation dataset on the baseline U-Net. Those same hyperparameters are then used for each of the models in our approach. Each model is trained using the Adam optimizer up to a maximum number of epochs and the best model with the best validation loss is saved during training. The validation loss is calculated as the Dice score coefficient (DSC) over the validation dataset at the same resolution as the input images. We do not upscale the outputs for the validation loss as we do for calculating the final metrics. Each model was trained using PyTorch 1.10 on an Nvidia GeForce RTX 3080 GPU. Where possible, we have fixed the random seed value to “2022”, but we have also run the experiments on two other random seeds and obtained similar results.
我们在三个独立的数据集上评估了我们的方法,这些数据集在第3.1节中详细描述,以下分别称为细胞、主动脉和息肉数据集。首先,对于每个数据集,我们训练了粗略分割的U-Net、Res-U-Net++和DeepLabv3+网络,使用各种下采样的输入分辨率。这些模型也被用作基线模型,以便与我们的方法进行比较。为了评估我们的方法,我们使用相同的数据集组合、架构和输入大小来训练细分割模型。总共训练了100多个神经网络来评估我们的方法,包括基线网络和训练在裁剪图像上的网络。每个网络从头开始使用下采样的数据集进行训练。然后,网络的输出被上采样到数据集的原始分辨率,并使用这些输出计算度量指标。我们使用留出的测试数据集来报告本节中的所有结果。基线模型用作使用我们方法的实验中的粗略分割网络。每个网络使用的超参数在表1中报告。为了提供模型性能的客观度量指标,所有超参数都是使用基线U-Net的验证数据集进行调整的。然后,对我们方法中的每个模型使用相同的超参数。每个模型使用Adam优化器训练,直到达到最大的迭代次数,并且在训练期间保存最佳的验证损失模型。验证损失计算为在与输入图像相同分辨率的验证数据集上的Dice分数系数(DSC)。我们不像计算最终度量指标时那样对验证损失的输出进行上采样。每个模型都是使用PyTorch 1.10在Nvidia GeForce RTX 3080 GPU上训练的。在可能的情况下,我们将随机种子值固定为“2022”,但我们也在其他两个随机种子上运行了实验并获得了类似的结果。
Conclusions
结论
Downscaling is a common source of segmentation errors in neural networks. In this paper, we present an approach to training neural networks that reduces downscaling by utilizing two neural networks and salient crops. We show how training a second neural network on cropped image regions can improve segmentation performance on small input sizes with few downsides. Our approach improves segmentation metrics on downscaled images across different modalities and image sizes, especially in terms of recall. We show that, while this approach increases inference time, it allows for training using much larger batch sizes while maintaining the same segmentation metrics. Note that the goal of our method is not to produce state-of-the-art segmentation results on high-resolution images. Instead, the goal is to allow training on heavily downscaled images without sacrificing segmentation performance. Our approach is a general preprocessing method and can be applied to a variety of different segmentation tasks regardless of the underlying architecture, so long as the two networks output a segmentation mask. In addition, the rough segmentation portion can be substituted with any method that produces a bounding box for each object. Aside from object detection methods, the bounding boxes could also be determined manually by an expert, as shown in [4]. While we did not evaluate our approach on 3D networks in this paper, there is nothing in our approach that is specific to 2D images. Our approach can be extended to 3D images by using 3D neural network architectures and 3D bounding boxes for crop regions. In addition, our approach could greatly benefit from using transfer learning, as the two networks use the same underlying architecture. It is possible that the results could be further improved with good transfer learning datasets as well as more complex training regimes such as contrastive learning [26]. During the development of this paper, we experimented with training the architecture end-to-end but failed to produce ways for the rough segmentation network to converge in a stable manner. We plan to explore this further in future work. We believe that this approach will allow future researchers to train standard semantic segmentation neural networks on downscaled versions of very high-resolution images without sacrificing segmentation performance. We also show that these results are general across a variety of biomedical images and thus can be applied to a very large number of problems in this space.
在神经网络中,下采样是导致分割错误的常见原因。在本文中,我们提出了一种训练神经网络的方法,通过利用两个神经网络和显著的裁剪来减少下采样。我们展示了如何通过在裁剪的图像区域上训练第二个神经网络,可以在小输入尺寸上提高分割性能,且几乎没有不利因素。我们的方法在不同模态和图像尺寸的下采样图像上改进了分割度量指标,特别是在召回率方面。我们展示了,尽管这种方法增加了推理时间,但它允许使用更大的批量大小进行训练,同时保持相同的分割度量指标。注意,我们方法的目标不是在高分辨率图像上产生最先进的分割结果。相反,目标是允许在严重下采样的图像上进行训练,而不牺牲分割性能。我们的方法是一种通用的预处理方法,可以应用于各种不同的分割任务,无论底层架构如何,只要两个网络输出分割掩码。此外,粗略分割部分可以被可以用任何产生每个对象边界框的方法替代粗略分割。除了对象检测方法外,边界框也可以由专家手动确定,如[4]所示。虽然我们在本文中没有评估我们方法在3D网络上的应用,但我们的方法并不特定于2D图像。我们的方法可以通过使用3D神经网络架构和3D边界框作为裁剪区域来扩展到3D图像。此外,我们的方法可以极大地受益于使用迁移学习,因为这两个网络使用相同的底层架构。有可能通过良好的迁移学习数据集以及更复杂的训练机制(如对比学习[26])进一步提高结果。在开发本文期间,我们尝试了端到端训练架构,但未能找到使粗略分割网络以稳定方式收敛的方法。我们计划在未来的工作中进一步探索这一点。我们相信,这种方法将允许未来的研究人员在不牺牲分割性能的情况下,对非常高分辨率图像的下采样版本训练标准的语义分割神经网络。我们还展示了这些结果在多种生物医学图像中是通用的,因此可以应用于这一领域的大量问题。
Figure
图
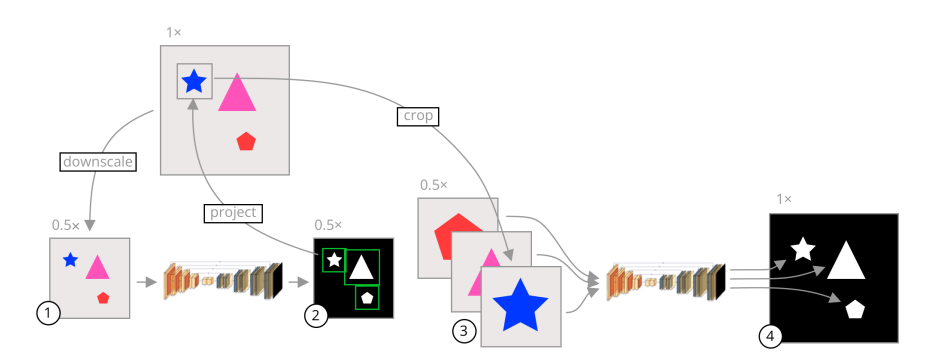
Figure 1. A visual summary of our approach. The gray images are the input images while the black images are segmentation mask outputs from the models. The shapes on the images are only representative, and the inputs can be any image where several objects need to be segmented. The arrows represent image operations. (1) An image is uniformly downsampled from its original resolution. (2) A rough segmentation is predicted by a neural network, and the bounding box of each connected component is calculated. (3) The bounding boxes are scaled to the original image space and crops of the input image are taken in the original resolution and scaled to a common input size. (4) Each crop is segmented separately by a second neural network specifically trained on cropped images. These crops are fused together to form a final segmentation in the original high resolution.
图 1. 我们方法的视觉总结。灰色图像是输入图像,而黑色图像是来自模型的分割掩码输出。图像上的形状仅代表性的,输入可以是任何需要分割多个对象的图像。箭头代表图像操作。(1) 图像从其原始分辨率均匀下采样。(2) 由神经网络预测大致分割,并计算每个连通部分的边界框。(3) 边界框被缩放到原始图像空间,并且以原始分辨率对输入图像进行裁剪并缩放到一个常见的输入大小。(4) 每个裁剪部分由专门针对裁剪图像训练的第二个神经网络单独分割。这些裁剪部分被融合在一起,形成原始高分辨率的最终分割。

Figure 2. The relationship between input dimensions and the mean Dice Score Coefficient (DSC) and recall for different datasets. The points are measured values from our experiments.
图2. 输入尺寸与不同数据集的平均Dice分数系数(DSC)和召回率之间的关系。点是我们实验中测量的值。
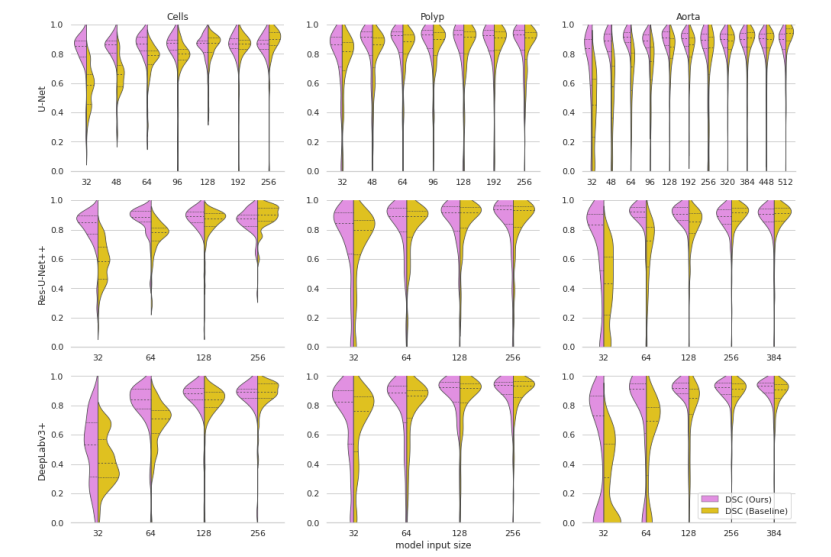
Figure 3. Violin plots of Dice Score Coefficients of our approach compared to the baseline models at various input dimensions. The dashed lines represent quartiles of the distributions.
图3. 我们方法与基线模型在不同输入尺寸下Dice分数系数的小提琴图。虚线代表分布的四分位数。

Figure 4. Example outputs from the models at various input sizes. (a) Cells dataset. (b) Polyp dataset. © Aorta dataset.
图4. 不同输入尺寸下模型的示例输出。(a) 细胞数据集。(b)息肉数据集。© 主动脉数据集。
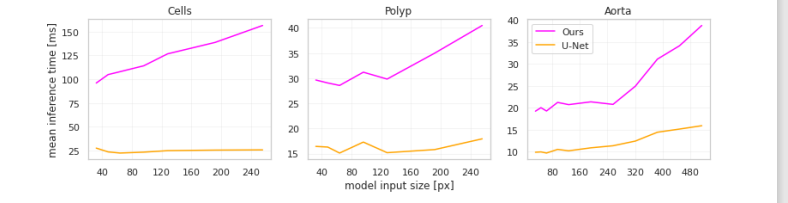
Figure 5. Mean per-input inference time across different input sizes for the U-Net-based models.
图5. 基于U-Net模型的不同输入尺寸下的平均每输入推理时间。
Table
表

Table 1. The hyper-parameters used for each of the models in our experiments.
表1. 我们实验中每个模型所使用的超参数。

Table 2. The results of our approach U-Net as the underlying architecture. The complete results of all of our experiments are available in Appendix A.
表2. 我们的方法使用U-Net作为基础架构的结果。我们所有实验的完整结果可在附录A中找到。

Table 3. A comparison of the Dice Score Coefficients of our approach using other underlying architectures at 4× and 2× downscaled images. The complete results of all of our experiments are available in Appendix A.
表3. 我们的方法在4倍和2倍下采样图像上使用其他基础架构时Dice分数系数的比较。我们所有实验的完整结果可在附录A中找到。
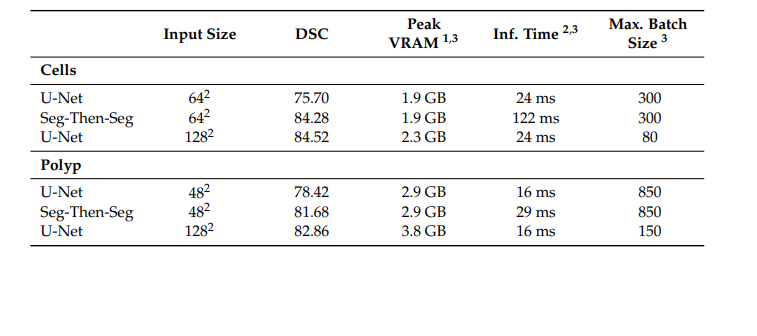

Table 4. Performance characteristics of our approach compared to the baseline model with similar mean test Dice Score Coefficients.
表4. 我们的方法与具有相似平均测试Dice分数系数的基线模型的性能特征比较。


Table A1. The full results of each of our U-Net experiments表A1.
我们每个U-Net实验的全部结果。

Table A2. The full results of each of our Res-U-Net++ experiment
表A2. 我们每个Res-U-Net++实验的全部结果。

Table A3. The full results of each of our DeepLabv3+ experiments
表A3. 我们每个DeepLabv3+实验的全部结果。