Title
题目
Nested resolution mesh-graph CNN for automated extraction of liver surfaceanatomical landmarks
用于肝脏表面解剖学标志自动提取的嵌套分辨率网格图卷积神经网络
01
文献速递介绍
基于增强现实(AR)的腹腔镜肝切除术导航引导 该技术引入了一种突破性的可视化方法,其核心是将术前三维肝脏模型(网格)与术中二维腹腔镜图像进行配准(Ali 等,2025;Lopez,2022;Pfeiffer 等,2018)。通过将肝脏模型叠加到腹腔镜视野中,外科医生能够准确识别内部结构(图 1A)。这种三维-二维融合的成功很大程度上依赖于将解剖学标志作为配准约束条件(Robu 等,2018;Mhiri 等,2024),例如肝嵴和镰状韧带。目前,这些解剖学标志在术前阶段需由人工在肝脏网格上标注(Koo 等,2017,2022)。尽管该过程在技术上可行,但耗时较长——每个病例通常需要数小时(Plantefeve 等,2016),且对解剖学专业知识要求极高。此外,在解剖形态各异的病例中实现标志的一致性识别仍面临挑战,尤其是涉及镰状韧带等细微标志时。这些局限性制约了 AR 辅助导航的可扩展性和标准化。因此,实现这些解剖学特征的自动提取(即分割)不仅是必要的,更是优化手术流程、推动 AR 技术更广泛临床应用的关键。 然而,从肝脏网格中自动分割关键解剖学标志具有挑战性,因为这需要全面理解全局几何结构(空间关系)和局部拓扑信息(网格单元形状)。如图 1A 所示,两种标志常用于视觉导航:镰状韧带和肝嵴。镰状韧带是一种薄纤维结构,连接肝脏前表面与腹壁,通常位于肝脏表面中线偏左区域,处于相对平坦、低曲率的区域,由大型、均匀分布的三角形面构成。但定义该结构的局部表面特征(如方向边缘对齐或细微曲率过渡)往往过于微弱,难以被人工标注者可靠识别。这促使研究人员开发能够学习这些具有解剖学意义但视觉上模糊模式的自动方法。相比之下,肝嵴位于肝脏前底部,横向延伸至两叶,处于高曲率区域,其特征为小型、密集聚集的三角形单元和更锐利的局部拓扑结构。 术前-术中腹腔镜融合(P2ILF)挑战赛(Ali 等,2025)是在肝图像术前-术中融合范围内解决该分割任务的开创性尝试。该挑战赛提供了包含 11 个肝脏网格的数据集,激发了全球研究界提出多种创新解决方案。参赛者提出的方法包括基于点的方法(如 Pointnet++(Qi 等,2017b))、基于图的方法(Kipf 和 Welling,2016)以及针对网格的策略(如 MeshCNN(Hanocka 等,2019),即获胜方法)。Pointnet++(Qi 等,2017b)和图卷积网络(GCN)方法(Kipf 和 Welling,2016)通过聚合点云特征在全局理解方面表现出色,但往往忽略了网格表面的丰富细节。相反,基于 MeshCNN 的方法(Hanocka 等,2019)通过网格卷积(MeshConv)沿网格表面拓扑结构进行特征计算,具备强大的局部拓扑学习能力。然而,在处理分辨率高且不一致的网格时(图 1B),由于其采用序列池化和边缘折叠策略,MeshCNN(Hanocka 等,2019)在计算效率和全局形状一致性表征方面存在问题。当处理具有不同肝脏形态和网格复杂度的样本中的解剖结构时,这些问题尤为突出(详见 3.2.1 节)。 为解决上述挑战,本文提出一种新颖的几何深度学习框架,该框架结合了基于图和基于网格方法的优势,同时弥补了它们各自的不足。具体而言,我们的嵌套分辨率网格-图卷积神经网络(Nested Resolution Mesh-Graph CNN)框架旨在准确提取关键肝脏解剖学标志(如镰状韧带和肝嵴),这些标志在三维肝脏网格上表现为顶点和边缘的组合。因此,该任务被重新表述为网格上的顶点或边缘分割问题。我们的方法在两个网格分辨率层级上运行:压缩的低分辨率网格和原始的高分辨率网格。对于低分辨率网格,我们利用动态图卷积网络(DGCNN)(Wang 等,2019b)快速学习肝脏的整体形状和外观,生成初始解剖学标志分割结果。通过不同的传播方法将这些分割结果映射到高分辨率网格表面,得到“扩张”(dilation)和“腐蚀”(erosion)层级的标志候选区域。随后,我们设计了一个基于 MeshConv 的解剖学优化网络,将这些标志候选区域与原始高分辨率网格的详细拓扑结构相结合。该网络采用基于细粒度聚合的注意力机制,有效平衡并融合来自不同候选区域的潜在正确先验信息,生成对肝脏表面复杂拓扑结构敏感的精细化分割结果。此外,我们引入了解剖学感知骰子损失(anatomy-aware Dice loss),该损失函数能够解决网格表面的不均匀性,并捕捉韧带和嵴的相对关系,进一步提升分割性能。 总之,本研究的主要贡献如下: 1. 提出一种嵌套分辨率框架,该框架整合动态图卷积(DGCNN)与基于 MeshConv 的优化模块,能够对不同分辨率的网格实现准确的肝脏标志分割。 2. 引入一种解剖学优化策略,通过注意力融合和辅助监督自适应整合粗粒度标志候选区域,并结合解剖学感知骰子损失缓解分辨率不平衡问题,提升分割质量。 3. 提供一个包含 200 个肝脏网格的人工标注数据集(源自公开 CT 数据集),并在临床相关的 P2ILF 挑战赛病例上验证该方法,证明其在不同成像模态和网格重建流程中的稳健泛化能力。 本文结构如下:第 2 节全面综述该领域的相关应用和研究工作;第 3 节详细描述数据和所提出的框架;第 4 节呈现实验设置和结果;最后,在第 5 节对本研究进行讨论和总结。
Aastract
摘要
The anatomical landmarks on the liver (mesh) surface, including the falciform ligament and liver ridge,are composed of triangular meshes of varying shapes, sizes, and positions, making them highly complex.Extracting and segmenting these landmarks is critical for augmented reality-based intraoperative navigationand monitoring. The key to this task lies in comprehensively understanding the overall geometric shape andlocal topological information of the liver mesh. However, due to the liver’s variations in shape and appearance,coupled with limited data, deep learning methods often struggle with automatic liver landmark segmentation.To address this, we propose a two-stage automatic framework combining mesh-CNN and graph-CNN. In thefirst stage, dynamic graph convolution (DGCNN) is employed on low-resolution meshes to achieve rapid globalunderstanding, generating initial landmark proposals at two levels, ‘‘dilation" and ‘‘erosion", and mapping themonto the original high-resolution surface. Subsequently, a refinement network based on mesh convolution fusesthese landmark proposals from edge features along the local topology of the high-resolution meshsurface,producing refined segmentation results. Additionally, we incorporate an anatomy-aware Dice loss to addressresolution imbalance and better handle sparse anatomical regions. Extensive experiments on two liver datasets,both in-distribution and out-of-distribution, demonstrate that our method accurately processes liver meshes ofdifferent resolutions, outperforming state-of-the-art methods.
肝脏(网格)表面的解剖学标志(包括镰状韧带和肝嵴)由形状、大小和位置各异的三角形网格构成,因此具有高度复杂性。这些标志的提取与分割对于基于增强现实的术中导航和监测至关重要。该任务的关键在于全面理解肝脏网格的整体几何形状和局部拓扑信息。然而,由于肝脏在形态和外观上存在个体差异,且数据量有限,深度学习方法在肝脏标志自动分割任务中往往面临挑战。 为解决这一问题,我们提出一种结合网格卷积神经网络(mesh-CNN)和图卷积神经网络(graph-CNN)的两阶段自动分割框架。第一阶段,在低分辨率网格上采用动态图卷积(DGCNN)实现快速全局特征理解,生成“扩张”(dilation)和“腐蚀”(erosion)两个层级的初始标志候选区域,并将其映射到原始高分辨率表面。随后,基于网格卷积的优化网络利用高分辨率网格表面的局部拓扑结构,融合来自边缘特征的标志候选区域,得到精细化分割结果。此外,我们引入解剖学感知骰子损失(anatomy-aware Dice loss),以解决分辨率不平衡问题,并更好地处理稀疏解剖区域。在两个肝脏数据集(分布内和分布外)上的大量实验表明,我们的方法能够准确处理不同分辨率的肝脏网格,性能优于当前主流方法。
Method
方法
3.1. Datasets
In this study, we manually annotated 200 liver meshes to evaluatethe proposed method and to promote further research in this field.These meshes were derived from three publicly available liver datasets:3Dircadb (Soler et al., 2010), LiTS (Bilic et al., 2023), and Amos (Jiet al., 2022). The surface models were first extracted from ground truthsegmentations using the Marching Cubes algorithm in 3D Slicer (Fedorov et al., 2012), and further processed using MeshLab (Cignoniet al., 2008) to ensure manifold simplification, compression, and watertightness. The falciform ligament and liver ridge were manuallyannotated as vertex-level regions on the liver mesh using Blendersoftware. Their annotation extents were defined with reference toanatomical landmarks: the falciform ligament extended upward fromthe midline fissure toward the superior surface, terminating approximately 1–1.5 cm below the highest point of the liver. The liver ridgeextended bilaterally from the fissure toward the lateral boundaries,ending approximately 3–5 cm inward from the endpoints of the liver’slongest transverse diameter. While the liver meshes underwent nearlyidentical processing steps, the resolution of the resulting meshes, measured in edge counts, varied significantly from 3000 to 20000 edges.This variation was not intentionally introduced but rather arose fromthe inherent differences in liver shapes and sizes across the datasets.These natural variations in liver morphology present a primary challenge in this task, highlighting the need for a robust segmentationapproach.All annotations were meticulously reviewed by clinical experts toensure accuracy and fairness in evaluation. Additionally, to furtherassess the generalizability of our proposed method, we included 9training cases from the P2ILF challenge (Ali et al., 2025) as an externalevaluation cohort. These meshes were generated through heterogeneous reconstruction pipelines (including CT and MRI modalities) withunknown meshing strategies, often resulting in distinct local surfacecharacteristics compared to our internal dataset.
3.1 数据集 本研究手动标注了200个肝脏网格,用于评估所提方法并推动该领域的进一步研究。这些网格来源于三个公开可用的肝脏数据集:3Dircadb(Soler 等,2010)、LiTS(Bilic 等,2023)和 Amos(Ji 等,2022)。首先利用 3D Slicer 软件中的移动立方体算法(Marching Cubes algorithm)(Fedorov 等,2012)从真实分割结果中提取表面模型,随后通过 MeshLab 软件(Cignoni 等,2008)进行后续处理,以确保网格的流形简化、压缩和密闭性。采用 Blender 软件在肝脏网格上以顶点级区域的形式手动标注镰状韧带和肝嵴,标注范围参考解剖学标志定义:镰状韧带从中线裂向上延伸至肝脏上表面,终止于肝最高点下方约 1-1.5 cm 处;肝嵴从裂向双侧延伸至外侧边界,终止于肝脏最长横径端点向内约 3-5 cm 处。尽管所有肝脏网格均经过近乎相同的处理步骤,但以边数衡量的网格分辨率差异显著(3000-20000 条边)。这种差异并非刻意引入,而是源于不同数据集间肝脏形态和大小的固有差异。肝脏形态的这些自然变异是该任务的主要挑战之一,也凸显了稳健分割方法的必要性。 所有标注均经临床专家仔细审核,以确保评估的准确性和公正性。此外,为进一步验证所提方法的泛化能力,我们将 P2ILF 挑战赛(Ali 等,2025)的 9 个训练病例纳入外部评估队列。这些网格通过异质性重建流程(包括 CT 和 MRI 模态)生成,其网格划分策略未知,与内部数据集相比,往往具有独特的局部表面特征。
Conclusion
结论
This study addresses the critical need for effective 3D landmarkextraction algorithms for AR-assisted laparoscopic navigation by developing a deep learning-based segmentation algorithm for two criticalanatomical landmarks on the liver surface: the falciform ligament andthe liver ridge. Our proposed algorithm employs a nested resolutionnetwork architecture that combines a Dynamic Graph CNN (DGCNN)and a MeshConv-based anatomical refining network to tackle the challenges posed by the significant variability in shape and appearanceof the liver surface. Initially, the liver mesh at its original resolution is pooled into a low-resolution mesh with a fixed number ofedges through a random edge collapse strategy, and the first-stagelandmark segmentation is generated by the DGCNN model. The landmark segmentation results at low resolution are then propagated tothe high-resolution mesh surface using the unpooling operation inMeshCNN, resulting in ‘‘dilation’’ and ‘‘erosion’’ landmark proposals.Subsequently, we design an anatomical refining network that integrates the landmark proposals, edge features from the high-resolutionmesh, and resolution encoding. This network, leveraging MeshConvbased specialized branches and an attention fusion module, extractsthe correct priors from the previous stage, ultimately achieving accurate landmark segmentation on original meshes of varying resolutions.Empirical evaluations on two liver mesh datasets demonstrate that ourframework consistently outperforms existing methods in terms of bothaccuracy and robustness.As hepatobiliary surgeries increasingly move toward minimally invasive and precision approaches, the design and development of ARnavigation systems for laparoscopic procedures have garnered significant attention. The 3D–2D registration (Sun, 2023) constrained byanatomical landmarks is foundational to realizing this groundbreakingvisualization technique. However, most related studies still rely onmanual annotation of key anatomical landmarks on 3D liver meshes bysurgeons, largely due to the considerable variability in liver appearanceand shape, as well as the lack of mesh data annotated with landmarks. This has presented significant challenges for the developmentof deep learning-based automated landmark segmentation methods.In response, we reconstructed and annotated 200 watertight meshesof varying shapes, appearances, and resolutions from three publiclyavailable liver CT datasets to develop and validate our liver landmarksegmentation algorithm. Additionally, we applied the proposed methodto the external P2ILF dataset collected from real patients for furthervalidation. The experimental results in Table 4 affirm the superiority ofour approach, particularly in the extraction of the falciform ligament,where it reduced the 3D Chamfer distance by approximately 24 mmcompared to DGCNN.
本研究针对增强现实(AR)辅助腹腔镜导航中高效三维标志提取算法的关键需求,提出了一种基于深度学习的分割算法,用于提取肝脏表面两个关键解剖学标志(镰状韧带和肝嵴)。所提算法采用嵌套分辨率网络架构,结合动态图卷积神经网络(DGCNN)与基于网格卷积(MeshConv)的解剖学优化网络,以应对肝脏表面形态和外观显著变异带来的挑战。首先,通过随机边缘折叠策略将原始分辨率的肝脏网格池化为固定边数的低分辨率网格,利用DGCNN模型生成第一阶段的标志分割结果;随后,借助MeshCNN中的反池化操作将低分辨率下的标志分割结果传播至高分辨率网格表面,得到“扩张”(dilation)和“腐蚀”(erosion)两级标志候选区域;最后,设计解剖学优化网络,整合标志候选区域、高分辨率网格的边缘特征及分辨率编码信息,该网络通过基于MeshConv的专用分支和注意力融合模块,从前期结果中提取正确先验信息,最终实现对不同分辨率原始网格的精准标志分割。在两个肝脏网格数据集上的实证评估表明,该框架在准确性和稳健性方面均持续优于现有方法。 随着肝胆外科手术日益向微创化和精准化方向发展,腹腔镜手术AR导航系统的设计与开发受到广泛关注。以解剖学标志为约束的三维-二维配准(Sun,2023)是实现这一突破性可视化技术的基础。然而,由于肝脏外观和形态存在显著个体差异,且缺乏带标志标注的网格数据,大多数相关研究仍依赖外科医生手动标注三维肝脏网格上的关键解剖学标志,这为基于深度学习的自动标志分割方法研发带来了巨大挑战。为此,本研究从三个公开肝脏CT数据集重建并标注了200个具有不同形态、外观和分辨率的密闭网格,用于开发和验证肝脏标志分割算法;同时,将所提方法应用于来自真实患者的外部P2ILF数据集进行进一步验证。表4的实验结果证实了该方法的优越性,尤其是在镰状韧带提取任务中,与DGCNN相比,三维倒角距离(3D Chamfer Distance)减少了约24毫米。
Results
结果
4.1. Competing methods and experimental setup
To thoroughly validate our method, we randomly divided the manually annotated liver mesh data into training, validation, and test sets ina 10:3:7 ratio, reporting the average performance on the test set acrossmultiple experiments.Currently, no publicly available algorithms specifically address thesegmentation of liver surface landmarks. Therefore, we selected fourstate-of-the-art (SOTA) deep learning methods for mesh data processing: Pointnet++ (point-based), DGCNN (graph-based), MeshCNN (edgebased), and TSGCN (designed for dental meshes). Pointnet++ andMeshCNN were employed by multiple teams in the P2ILF challenge (Aliet al., 2025), demonstrating their capability on liver meshes. DGCNN,known for its strong local and global learning abilities, was also chosenfor processing the compressed low-resolution meshes in our study.Additionally, the original MeshCNN, which takes five initial edge features as input, excels in local understanding but struggles with globalinformation. To mitigate this limitation, we computed the center pointcoordinates of the edges as an additional input, enhancing MeshCNNand designating this variant as MeshCNN-1 for comparison. Furthermore, TSGCN, which incorporates dual parallel graph networks andleverages attention mechanisms for feature fusion, is specialized fordental mesh segmentation tasks, offering an additional perspective inour comparative analysis.All methods, including ours, were implemented in PyTorch andexecuted on an RTX 8000 GPU. For Pointnet++ and DGCNN, the training involved random sampling of spatial coordinates (size 3000 × 3)while testing inputs on all edges to obtain segmentation results onthe original resolution mesh. MeshCNN and MeshCNN-1 employed aUNet-like architecture with three levels of down-sampling, processingfixed-size input data of edges (size 20000 × 5). We also used the officialimplementation of TSGCN for training and evaluation, modifying thenumber of input edges to 3000, matching the DGCNN setting. Finally,in our method, we first obtained different levels of landmark proposalsusing a trained DGCNN model, followed by training the anatomicalrefining network with original meshes at varying resolutions. All experiments were optimized using the Adam optimizer with a learningrate of 0.01, and models with a minimum loss of over 600 epochs wereselected for testing
4.1 对比方法与实验设置 为全面验证所提方法的性能,我们将手动标注的肝脏网格数据按 10:3:7 的比例随机划分为训练集、验证集和测试集,并报告多次实验在测试集上的平均性能。 目前尚无专门针对肝脏表面标志分割的公开算法,因此我们选取了四种处理网格数据的当前主流(SOTA)深度学习方法进行对比:Pointnet++(基于点)、DGCNN(基于图)、MeshCNN(基于边)以及 TSGCN(专为牙齿网格设计)。其中,Pointnet++ 和 MeshCNN 已被多个团队应用于 P2ILF 挑战赛(Ali 等,2025),证明了其在肝脏网格上的有效性;DGCNN 因其强大的局部和全局学习能力,被选为本文中低分辨率压缩网格的处理方法。此外,原始 MeshCNN 以 5 维初始边缘特征作为输入,在局部特征理解方面表现出色,但在全局信息捕捉上存在不足。为缓解这一局限性,我们计算了边缘的中心点坐标作为额外输入对其进行改进,并将该变体命名为 MeshCNN-1 用于对比实验。另外,TSGCN 融合了双并行图网络并利用注意力机制进行特征融合,专为牙齿网格分割任务设计,可为本次对比分析提供额外视角。 所有方法(包括本文方法)均基于 PyTorch 框架实现,并在 RTX 8000 GPU 上运行。对于 Pointnet++ 和 DGCNN,训练过程中随机采样 3000×3 维度的空间坐标数据,测试时则输入所有边缘以获取原始分辨率网格上的分割结果;MeshCNN 和 MeshCNN-1 采用类 UNet 架构,包含三级下采样,处理 20000×5 维度的固定尺寸边缘输入数据;TSGCN 采用官方实现进行训练和评估,仅将输入边缘数量修改为 3000,与 DGCNN 的设置保持一致。本文方法的实验流程为:首先利用训练好的 DGCNN 模型生成不同层级的标志候选区域,随后使用不同分辨率的原始网格训练解剖学优化网络。所有实验均采用 Adam 优化器(学习率设为 0.01)进行优化,最终选取训练 600 个 epoch 以上且损失值最小的模型用于测试。
Figure
图

Fig. 1. (A) Accurate segmentation of anatomical landmarks on the liver surface is crucial for developing intraoperative AR navigation systems. The magnifiedinset highlights landmark vertices and their shared edges, which define the segmentation regions studied in this work. (B) The liver mesh surface lacks textureand exhibits significant variations in shape and appearance, making landmark segmentation particularly challenging, especially for the falciform ligament
图 1 (A) 肝脏表面解剖学标志的精准分割是开发术中AR导航系统的关键。放大插图突出显示了标志顶点及其共享边缘,这些顶点和边缘定义了本研究中的分割区域。(B) 肝脏网格表面缺乏纹理,且在形态和外观上存在显著差异,这使得标志分割极具挑战性,尤其是对于镰状韧带而言。
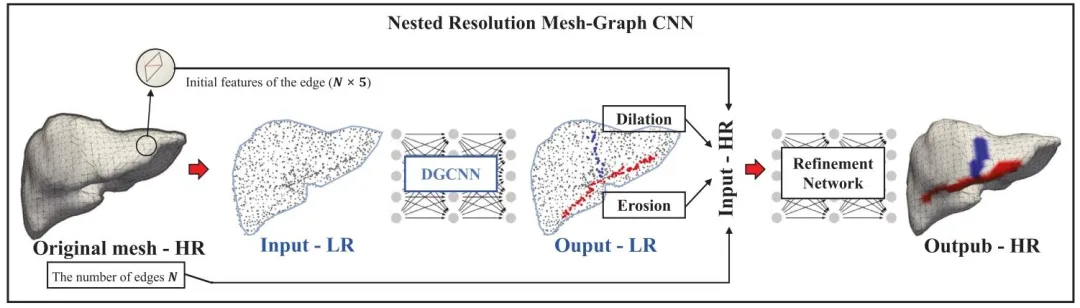
Fig. 2. The overall framework of our proposed nested resolution Mesh-Graph CNN for segmenting key anatomical landmarks on the surface of the liver, i.e. thefalciform ligament (in blue) and liver ridge (in red). Here, both sparse representation (denoted as LR for low resolution) and dense representation (denoted asHR for high resolution) of the point cloud are used. 𝑁 represents the number of edges in the original mesh
图 2 本文提出的嵌套分辨率网格-图卷积神经网络(Mesh-Graph CNN)的整体框架,用于分割肝脏表面关键解剖学标志(即蓝色标注的镰状韧带和红色标注的肝嵴)。该框架同时采用了点云的稀疏表示(记为低分辨率LR)和密集表示(记为高分辨率HR)。其中,𝑁 代表原始网格中的边数。

Fig. 3. (A) A close-up view of the single-ring structure of the liver surface and the input edge features, where 𝑎, 𝑏, 𝑐, 𝑑, 𝑒 represent the edges in the single-ringstructure. (B) Mesh convolution and the process of pooling and unpooling, resulting in new edges 𝑎 ′ , 𝑏′ , 𝑐′ , 𝑑′ , 𝑒′ . © The feature propagation process duringunpooling in MeshCNN. Here, 𝐹 ∈ R𝐻×𝐶 represents the feature matrix of the low-resolution mesh (with 𝐻 edges). Using the Unroll matrix 𝑀 ∈ R𝐻×𝑁 , thesefeatures are propagated to the high-resolution mesh level, yielding the corresponding features 𝐹 ′ ∈ R𝑁×𝐶 (with 𝑁 edges, where 𝑁 > 𝐻).
图 3 (A) 肝脏表面单环结构及输入边缘特征的特写视图,其中𝑎、𝑏、𝑐、𝑑、𝑒 代表单环结构中的边。(B) 网格卷积及池化与反池化过程,最终生成新边𝑎′、𝑏′、𝑐′、𝑑′、𝑒′。© MeshCNN 中反池化阶段的特征传播过程。此处,𝐹 ∈ R𝐻×𝐶 表示低分辨率网格的特征矩阵(含𝐻条边);通过展开矩阵(Unroll matrix)𝑀 ∈ R𝐻×𝑁,将这些特征传播至高分辨率网格层级,得到对应的特征矩阵𝐹′ ∈ R𝑁×𝐶(含𝑁条边,其中𝑁 > 𝐻)。 ### 关键术语说明(贴合学术规范) 1. 单环结构(single-ring structure):指肝脏网格表面由相邻边构成的环形局部拓扑结构,为网格卷积的核心作用区域; 2. 池化与反池化(pooling and unpooling):深度学习中网格数据的特征降维与升维操作,此处保留计算机图形学领域标准译法; 3. 展开矩阵(Unroll matrix):MeshCNN 中用于低分辨率特征向高分辨率网格映射的专用矩阵,采用领域惯用命名直译; 4. 特征矩阵(feature matrix):维度表示遵循数学规范(R行×列),𝐻(低分辨率边数)、𝑁(高分辨率边数)、𝐶(特征通道数)为论文定义变量,保留原符号表述。
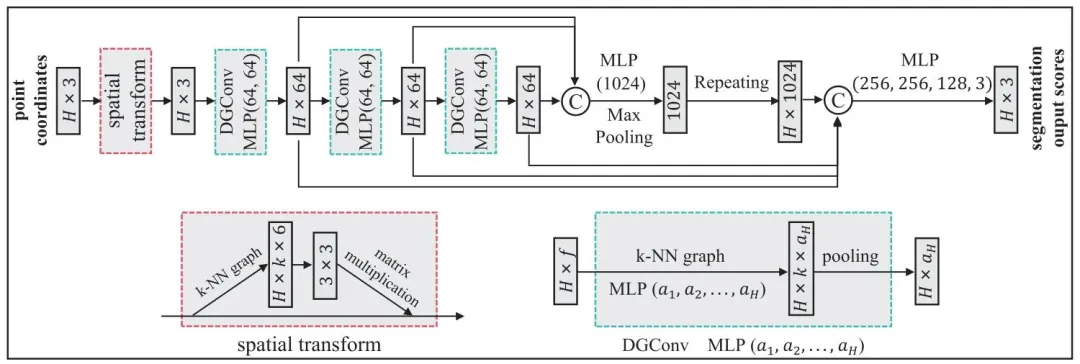
Fig. 4. Dynamic Graph Convolutional Neural Network (DGCNN) architecture used for the initial segmentation of anatomical landmarks on compressed lowresolution meshes. The network includes a spatial transform module for normalizing input edge features, followed by dynamic graph convolution (DGConv)layers that extract multi-level features. Finally, Multi-Layer Perceptron (MLP) layers aggregate these features to produce initial segmentation scores. 𝑘-NearestNeighborhood (𝑘-NN) is used in both spatial and DGConv layers
图 4 用于压缩低分辨率网格上解剖学标志初始分割的动态图卷积神经网络(DGCNN)架构。该网络包含一个用于归一化输入边缘特征的空间变换模块,随后是提取多级特征的动态图卷积(DGConv)层。最后,多层感知机(MLP)层对这些特征进行聚合,生成初始分割分数。空间变换层和动态图卷积(DGConv)层均采用𝑘近邻(𝑘-NN)算法。 ### 术语规范说明 1. 空间变换模块(spatial transform module):深度学习中用于特征空间归一化的标准模块,采用领域通用译法; 2. 动态图卷积层(DGConv layers):动态图卷积神经网络(DGCNN)的核心组件,保留模型缩写+层(layers)的规范表述; 3. 多层感知机(MLP):深度学习基础模型,采用领域通用缩写及全称对应译法; 4. 𝑘近邻(𝑘-NN):机器学习经典算法,保留原符号𝑘及缩写NN(Nearest Neighborhood)的标准译法,括号内补充完整英文表述。
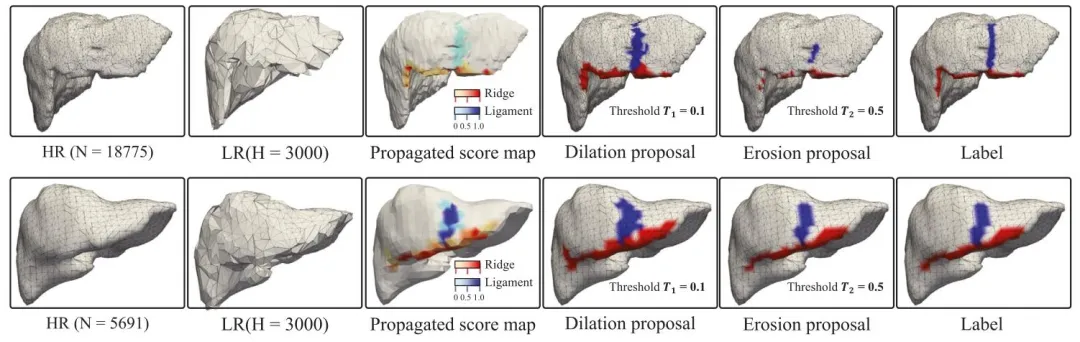
Fig. 5. Comparison with two original meshes with different resolutions (from left to right): the appearance of the (original) high-resolution mesh (HR), theappearance of the compressed low-resolution (LR) mesh, the propagation score map (PSM) mapping the initial segmentation onto the surface of the original highresolution mesh, the ‘‘dilation’’ landmark proposal based on threshold 𝑇1 , the ‘‘erosion’’ landmark proposal based on threshold 𝑇2 , and the landmark annotationon the original high-resolution mesh referred as ‘‘label’’.
图 5 两种不同分辨率原始网格的对比结果(从左至右):(原始)高分辨率网格(HR)外观、压缩低分辨率网格(LR)外观、将初始分割结果映射至原始高分辨率网格表面的传播分数图(PSM)、基于阈值𝑇₁的“扩张”(dilation)标志候选区域、基于阈值𝑇₂的“腐蚀”(erosion)标志候选区域,以及原始高分辨率网格上的标志标注(记为“label”)。

Fig. 6. (A) Architecture of the anatomical refining network based on MeshConv. Auxiliary segmentation heads (ASH) promote reliable prior extraction, whilethe attention fusion module (AFM) integrates these priors into refined landmark predictions. The first MeshConv (in red) omits symmetric functions (i.e., Eq. (2))due to resolution-encoded inputs. (B) Attention fusion module (AFM) based on fine-grained aggregation (FGA), where superscripts (1) and (2) indicate separateinstances used to enhance cross-branch communication. © Edge weights incorporated in our anatomy-aware Dice (AAD) losses
图 6 (A) 基于网格卷积(MeshConv)的解剖学优化网络架构。辅助分割头(ASH)用于促进可靠先验信息的提取,而注意力融合模块(AFM)则将这些先验信息整合到精细化的标志预测结果中。第一个网格卷积层(红色标注)由于输入包含分辨率编码信息,因此省略了对称函数(即公式(2))。(B) 基于细粒度聚合(FGA)的注意力融合模块(AFM),其中上标(1)和(2)表示用于增强跨分支通信的独立实例。© 融入解剖学感知骰子损失(AAD)的边缘权重示意图。
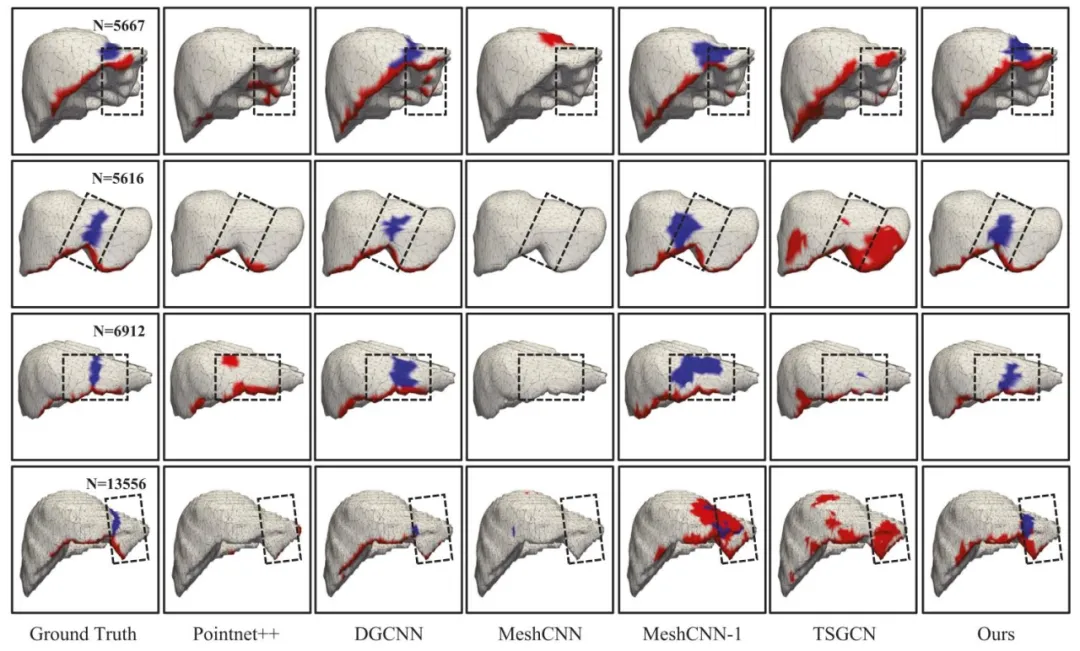
Fig. 7. Comparison of segmentation results of different segmentation methods. The falciform ligament on the surface of the liver is shown in blue, and the liverridge is shown in red. In addition, the dotted boxes in the figure indicate areas to focus on.
图 7 不同分割方法的分割结果对比。肝脏表面的镰状韧带以蓝色显示,肝嵴以红色显示。图中虚线框标注为重点关注区域。

Fig. 8. Visual analysis of the attention fusion module (AFM) and auxiliary segmentation heads (ASH). Three representative liver meshes with varying resolutionsare shown in the first column, overlaid with ground truth landmarks. The next two columns show coarse landmark proposals obtained by applying differentconfidence thresholds (𝑇1 < 𝑇2 ) to the propagated score maps. AFM columns visualize attention weights from both branches, while ASH outputs illustrate howauxiliary supervision improves proposal quality. The final column shows refined predictions after integration.
图 8 注意力融合模块(AFM)与辅助分割头(ASH)的可视化分析。第一列展示三个不同分辨率的代表性肝脏网格,叠加了真实标志标注(ground truth landmarks);接下来两列显示对传播分数图(propagated score maps)应用不同置信度阈值(𝑇₁ < 𝑇₂)得到的粗粒度标志候选区域;AFM 列可视化了两个分支的注意力权重,ASH 输出列展示了辅助监督如何提升候选区域质量;最后一列显示整合后的精细化预测结果。
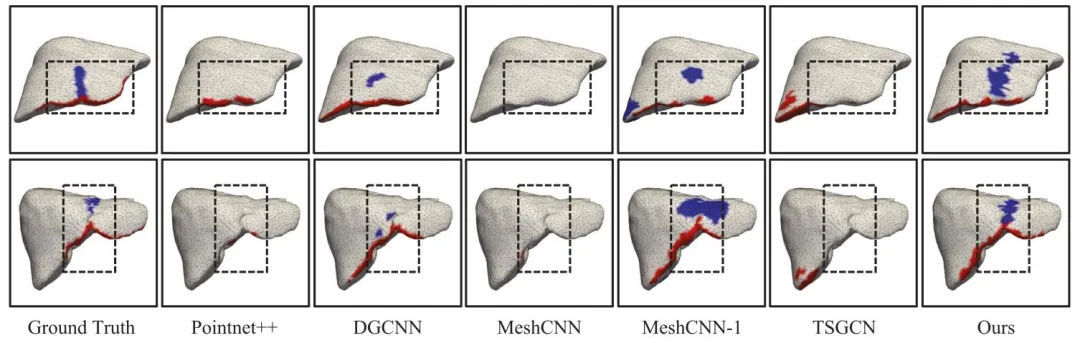
Fig. 9. Generalizability analysis on external dataset — visual results from P2ILF challenge dataset. In addition, the dotted boxes in the figure indicate areas tofocus on
图 9 外部数据集上的泛化性分析——P2ILF挑战赛数据集的可视化结果。图中虚线框标注为重点关注区域。
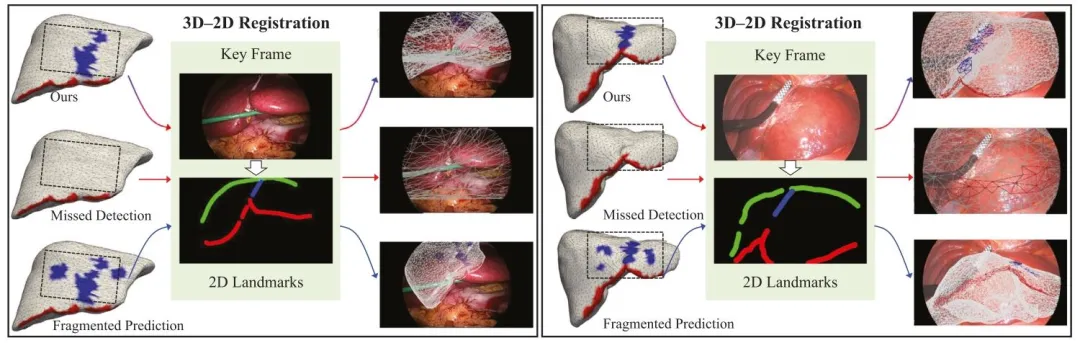
Fig. 10. Visual comparison of 3D–2D registration outcomes across three types of landmark segmentations on two P2ILF cases. Our method yields more completeand spatially coherent predictions for both ligament and ridge, enabling successful overlay, while the other two settings – with missing or fragmented landmarkregions – lead to failed or unstable alignment. The green curve indicates the extracted liver silhouette from the laparoscopic key frame. Registration was performedusing our PyTorch3D-basedpipeline. Supplementary videos demonstrate the dynamic rigid alignment process
图 10 两种P2ILF病例中三种标志分割结果的三维-二维配准效果可视化对比。我们的方法对韧带和嵴均能生成更完整、空间一致性更强的预测结果,从而实现成功叠加;而另外两种设置(标志区域缺失或破碎)则导致配准失败或不稳定。绿色曲线表示从腹腔镜关键帧中提取的肝脏轮廓。配准过程通过基于PyTorch3D的流程实现。补充视频展示了动态刚性配准过程。
Table
表
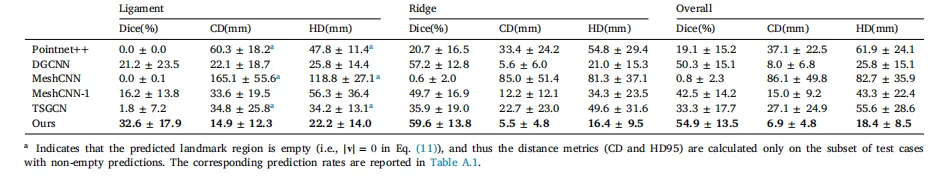
Table 1Quantitative comparison with different segmentation methods. ‘‘Overall’’ refers to the combined evaluation of both anatomical landmarks – falciform ligamentand liver ridge – as a single class, serving as a complementary summary metric
表 1 不同分割方法的定量对比。“总体(Overall)”指将镰状韧带和肝嵴这两个解剖学标志合并为单一类别进行综合评估,作为补充性汇总指标。

Table 2Ablation study on different key components. Wilcoxon signed-rank test and effect size (𝑟) were applied to the last four variants to evaluate statistical significanceand effect magnitude.
表 2 不同关键组件的消融实验结果。对最后四种变体采用威尔科克森符号秩检验(Wilcoxon signed-rank test)和效应量(𝑟)分析,以评估统计显著性和效应大小。
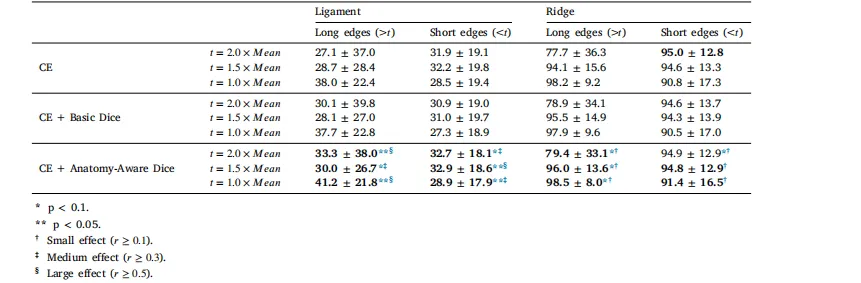
Table 3Dice value comparison of different loss function setups for Falciform Ligament and Liver Ridge, considering edge lengthdistributions. The threshold 𝑡 is defined as a multiple of the mean edge length (𝑀𝑒𝑎𝑛), where edges longer than 𝑡 are considered‘‘Long Edges’’ (which are sparse and important on the liver surface), and edges shorter than 𝑡 are ‘‘Short Edges’’ (which are denserand more frequent). The last three rows (using Anatomy-Aware Dice) were further evaluated using Wilcoxon signed-rank testsand effect size (𝑟) to assess significance and effect strength
表 3 考虑边缘长度分布时,不同损失函数设置下镰状韧带和肝嵴的骰子系数(Dice)对比。阈值𝑡定义为平均边缘长度(𝑀𝑒𝑎𝑛)的倍数,其中长度大于𝑡的边缘被视为 “长边缘”(在肝脏表面稀疏且重要),长度小于𝑡的边缘被视为 “短边缘”(更密集且出现频率更高)。最后三行(采用解剖学感知骰子损失(Anatomy-Aware Dice))通过威尔科克森符号秩检验(Wilcoxon signed-rank test)和效应量(𝑟)进一步评估,以验证统计显著性和效应强度。

Table 4External test results on P2ILF dataset — 3D Chamfer Distance (CD).Ligament Ridge Overall
表 4 P2ILF 数据集上的外部测试结果——三维倒角距离(3D Chamfer Distance, CD)。 (列名:韧带、嵴、总体)

Table 5Efficiency comparison of segmentation methods in terms of Parameters and Inference Time per sample.
表 5 不同分割方法在参数数量(Parameters)和单样本推理时间(Inference Time per sample)方面的效率对比。
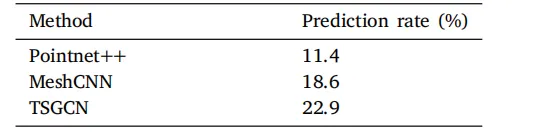
Table A.1Prediction rates (%) for the falciform ligamentlandmark on the internal test set (Number of testcases = 70). Prediction rate is defined as the percentage of test cases with non-empty predictions.
表 A.1 内部测试集上镰状韧带标志的预测率(%)(测试病例数 = 70)。预测率定义为预测结果非空的测试病例占比。

Table A.2Prediction rates (%) for the falciform ligamentlandmark on the P2ILF data (9 cases used forexternal evaluation). Prediction rate is definedas the percentage of test cases with non-emptypredictions.
表 A.2 P2ILF 数据集上镰状韧带标志的预测率(%)(用于外部评估的病例数 = 9)。预测率定义为预测结果非空的测试病例占比。