Title
题目
Re-identification from histopathology images
组织病理学图像的再识别
01
文献速递介绍
在光学显微镜下评估苏木精和伊红(H&E)染色切片是肿瘤病理诊断的标准程序。随着全切片扫描仪的出现,能够将玻璃切片数字化为所谓的全切片图像(WSI)。这一技术使得肿瘤评估自动化方法的使用成为可能,特别是深度学习(DL)算法的应用,彻底改变了数字病理学领域。这些模型在多种任务中表现出了令人鼓舞的性能,例如自动肿瘤分级和分类(Nir等,2018;Han等,2017;Ganz等,2021),自动生物标志物评估(如有丝分裂计数,Aubreville等,2023;Veta等,2019),以及肿瘤区域的分割(Wilm等,2022)。一些DL算法甚至能够从WSI中提取出人类专家无法察觉的信息,例如预测分子改变(Coudray等,2018;Hong等,2021;Lu等,2021b)或预测转移癌的原发部位(Lu等,2021a)。
大多数DL方法的共同点是需要大量数据进行训练。这促使了大规模组织病理学数据集的发布以加速DL研究,例如CAMELYON数据集(Litjens等,2018)、癌症基因组图谱(The Cancer Genome Atlas, TCGA)的乳腺浸润性癌数据集(Lingle等,2016)以及肿瘤增殖评估挑战2016数据集(Veta等,2019)。
在临床实践中,WSI通常伴随有患者的私人信息,例如姓名、年龄、性别等。这些信息被归类为受保护的健康信息(PHI),在大多数国家受到政府法规的保护,例如美国的《健康保险携便与责任法案》(HIPAA,Centers for Disease Control and Prevention, 2023)或欧洲的《通用数据保护条例》(GDPR,European Union, 2023)。因此,数据匿名化是发布医学数据过程中的关键步骤(Willemink等,2020;Moore等,2015;Bisson等,2023)。在WSI的背景下,这意味着需要移除所有可能存在PHI的地方,例如文件名或任何切片标签。此外,还需要仔细检查切片文件的元数据(Clark等,2013)。
如果图像中包含可用作生物特征标识符的信息,例如视网膜扫描或指纹,则可能出现潜在问题,因为这些信息可以作为“身份签名”,被视为PHI(Willemink等,2020)。虽然指纹和视网膜扫描通常被公认为生物特征标识符,其他临床数据(如头颈部CT扫描)也可能包含高度患者特异性的信息。例如,头颈部CT扫描的软组织重建可以用于还原患者面部以进行身份识别(Willemink等,2020)。
鉴于DL算法从图像中提取语义特征的能力,以前未被认为是“身份签名”的医学图像数据现在看起来足够患者特异性,可以用来重新识别患者。例如,Packhäuser等(2022)表明,一个训练良好的DL算法可以从胸部正位X光片中重新识别患者,即使这些记录间隔了多年。
鉴于DL模型识别组织病理学切片中复杂模式的能力及其重新识别医学图像中患者的潜力,一个问题随之而来:WSI是否包含足够的患者特异性信息,使DL模型能够用于患者重新识别。如果存在这样的模型,将引发隐私问题,因为理论上存在两种可能的PHI泄露方式:第一种场景是,如果攻击者获得了一个未匿名化的切片,这种模型可以将该图像与匿名的公共数据集中存在的患者关联起来(Packhäuser等,2022;Esmeral和Uhl,2022)。如果这些匿名图像还关联有疾病、治疗或性别等元数据,甚至更多的PHI可能会泄露。第二种场景是,来自同一患者的图像分散在不同数据集中。这在罕见肿瘤或高级别肿瘤的情况下并非不可能,因为这些病例的患病率较低。在这种情况下,DL模型可以将所有这些图像分配给同一患者。如果以前匿名化的图像还关联有其他元数据,将这些数据合并可能提供足够的信息重新识别患者。鉴于上述潜在问题,本研究探讨是否可以利用DL算法在大型匿名化组织病理学数据集中进行患者重新识别。我们的研究由以下问题引导:R1:从组织病理学全切片图像中重新识别患者是否普遍可行?R2:在采样时间间隔较大的情况下,是否可以重新识别同一患者的切片?我们的贡献如下:我们设计了针对这些研究问题的实验,这是组织病理学领域首次对此类问题的探讨。我们确定了导致匿名化组织病理学样本成功重新识别的关键因素。我们制定了如何安全发布此类图像的指南,因为医学数据的发布对现代算法的开发至关重要。
Aastract
摘要
In numerous studies, deep learning algorithms have proven their potential for the analysis of histopathologyimages, for example, for revealing the subtypes of tumors or the primary origin of metastases. These modelsrequire large datasets for training, which must be anonymized to prevent possible patient identity leaks.This study demonstrates that even relatively simple deep learning algorithms can re-identify patients in largehistopathology datasets with substantial accuracy. In addition, we compared a comprehensive set of state-ofthe-art whole slide image classifiers and feature extractors for the given task. We evaluated our algorithms ontwo TCIA datasets including lung squamous cell carcinoma (LSCC) and lung adenocarcinoma (LUAD). We alsodemonstrate the algorithm’s performance on an in-house dataset of meningioma tissue. We predicted the sourcepatient of a slide with 𝐹1 scores of up to 80.1% and 77.19% on the LSCC and LUAD datasets, respectively,and with 77.09% on our meningioma dataset. Based on our findings, we formulated a risk assessment schemeto estimate the risk to the patient’s privacy prior to publication.
在众多研究中,深度学习算法已被证明在组织病理学图像分析方面具有潜力,例如用于揭示肿瘤亚型或转移癌的原发部位。这些模型需要大型数据集进行训练,并且必须匿名化以防止可能的患者身份泄露。本研究表明,即使是相对简单的深度学习算法,也能够以较高的准确率在大型组织病理学数据集中重新识别患者。此外,我们针对这一任务比较了一组全面的最先进全切片图像分类器和特征提取器。我们在两个TCIA数据集上评估了我们的算法,这两个数据集分别是肺鳞状细胞癌(LSCC)和肺腺癌(LUAD)。我们还在自有的脑膜瘤组织数据集上展示了算法的性能。研究中,我们预测了切片来源患者的身份,在LSCC和LUAD数据集上的F1分数分别达到了80.1%和77.19%,而在脑膜瘤数据集上的F1分数为77.09%。基于我们的研究结果,我们制定了一套风险评估方案,用于在数据集发布前评估患者隐私泄露的风险。
Method
方法
In this study, we utilized three distinct datasets of which two arepublicly available. Those two datasets, namely lung adenocarcinoma(LUAD) (National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), 2018a) and lung squamous cell carcinoma(LSCC) (National Cancer Institute Clinical Proteomic Tumor AnalysisConsortium (CPTAC), 2018b), were obtained from TCIA (Clark et al.,2013). In the remainder of this paper, these datasets will be referred toas the LUAD dataset and the LSCC dataset. These datasets were scannedat a resolution of 0.5 μm per pixel and were obtained from variouspathology centers. We restricted our analysis to slides of patients forwhich at least two slides were available, resulting in 1059 images of226 patients for the LUAD dataset and 1071 images of 209 patients ofthe LSCC dataset.
在本研究中,我们使用了三个不同的数据集,其中两个是公开可用的。这两个公开数据集分别是肺腺癌(LUAD)(National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), 2018a)和肺鳞状细胞癌(LSCC)(National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), 2018b),它们来自TCIA(Clark等,2013)。在本文的后续部分,这两个数据集将分别称为LUAD数据集和LSCC数据集。这些数据集以每像素0.5 μm的分辨率进行扫描,来自多个病理中心。我们将分析范围限制在至少包含两张切片的患者,最终LUAD数据集包含226名患者的1059张图像,LSCC数据集包含209名患者的1071张图像。
Conclusion
结论
This work demonstrates that re-identification of patients fromhistopathology images of resected tumor specimens is possible, withsome limitations. As long as the slides originate from the same tumor,we can re-identify the patients with considerable accuracy (as canbe seen in Tables 1 and 2). If the slides were resected at differentpoints in time, the accuracy is considerably lower (see Table 3). Asuccessful resection should completely remove the tumor, and hencea later resection resembles a regrowth of an incomplete resection or anew tumor of potentially different pathogenesis and mutational pattern.Our results indicate that the strong performance drop could be linkedto different morphological tumor characteristics. Consequently, ourapproach is more likely to identify tumors than patients.Which visual factors in particular contribute to the re-identificationis a question for future research. However, even if the models wouldheavily rely on traces related to slide preparation to re-identify theslides, this would threaten patient privacy. Therefore if these factorswould imprint some kind of implicit visual time stamp, future workcan focus on how to remove these traces from the slides.Our results indicate that the safest way of publishing histopathologyimages is to only use each patient in one data publication, as tracingacross datasets and hence recombination of multiple meta and imagedatasets is feasible, especially if slides originating from the same tumorare used in different datasets.
本研究表明,从切除肿瘤标本的组织病理学图像中重新识别患者是可能的,但存在一些限制。只要切片来源于同一肿瘤,我们便能够以相当高的准确率重新识别患者(见表1和表2)。然而,如果切片来自不同时间点的切除手术,准确率则显著降低(见表3)。一次成功的切除手术应完全清除肿瘤,因此后续的切除可能反映为不完全切除导致的肿瘤再生长,或可能具有不同病理特征和突变模式的新肿瘤。我们的结果表明,性能的大幅下降可能与肿瘤形态学特征的差异相关。因此,我们的方法更可能是在识别肿瘤而非患者。
具体哪些视觉因素对重新识别的贡献最大,是未来研究需要回答的问题。然而,即使模型主要依赖与切片制备相关的痕迹来重新识别切片,这也会威胁患者隐私。如果这些因素为切片隐含地添加了某种视觉时间戳,未来的工作可以重点研究如何从切片中移除这些痕迹。
我们的结果表明,发布组织病理学图像最安全的方法是确保每位患者仅在一次数据发布中使用。尤其是如果来自同一肿瘤的切片被用于不同的数据集中,那么跨数据集的追踪以及多组元数据和图像数据集的重组是可行的,这对患者隐私构成潜在威胁。
Results
结果
For all experiments, we report the recall@1, recall@5, the precisionand the 𝐹1 score. These values are always the average values over allclasses. When considering recall@n, it means that for an algorithm’spredictions to be considered correct, the searched patient has to beincluded among the 𝑛 patients with the highest-ranked predictionsbased on the classification score. In a multiclass classification problem,the average recall@1 equals the balanced accuracy. For comparison,the probability of selecting the right patient by chance when assessing𝑁 patients is also given for each dataset.
5.1. Results of experiment 1On all three datasets, the methods demonstrated satisfactory performance in re-identifying the patients based on histology slides. Detailedresults can be found in Table 1. On the MEN dataset, the TransMILclassifier in conjunction with the UNI feature extractor achieved thehighest recall@1 with 77.09%. The comparison of different featureencoders for projecting WSI-patches into latent space revealed thatthe UNI model yielded the most favorable results across all comparedWSI-classifiers. In general, each of the feature extractors pre-trainedon histopathology data demonstrated superior performance comparedto the ImageNet baseline. We also found satisfactory performances onthe two public datasets. Using the TransMIL approach with the UNIfeature extractor, we achieved a recall@1 of 77.19% on the LUAD andof 80.01% on the LSCC dataset.The results of Experiment 1 with stain augmentation are presentedin Table 2. In comparison to the results without augmentation presented in Table 1, the performances of all compared models decreasedwith the use of stain augmentation. The drop was most pronounced forthe patch-based and naive-MIL approaches, with recall@1 decreasingfrom 74.92% to 65.25% and from 74.98% to 61.13%, respectively. Incontrast, the drop in performance was less pronounced for the WSIclassifiers. Of these, the CLAM model exhibited the greatest drop inrecall@1, with a decrease from 66.54% to 62.97%, while the TransMILapproach yielded a constantly high recall@1 value of 76.14%.
5.2. Results of experiment 2When the models were trained on the earliest resection and testedon later resections of the MEN dataset, the performance dropped remarkably compared to Experiment 1 (see Table 3). The highest performance among the compared methods was observed for the TransMILmethod, with a recall@1 of 15.10% and a recall@5 of 29.25%. Eventhough the individual results were lower than in Experiment 1, theyall remained considerably above the respective probabilities of randomguessing.
5.3. Results of the post hoc analysis of experiments 1 and 2Fig. 6 illustrates the L2-distance between latent space embeddingsof test samples and their corresponding latent space anchors for Experiments 1 and 2. In both experiments, the correctly classified sampleswere observed to have a considerably smaller distance to their respective latent space anchors than the misclassified samples. Additionally,the distances between test samples and their latent space anchors wereconsiderably smaller in Experiment 1 than in Experiment 2.
在所有实验中,我们报告了 recall@1、recall@5、precision(精确率) 和 𝐹1 分数。这些指标均为各类别的平均值。对于 recall@n,其定义为算法的预测被认为正确的条件是目标患者需包含在基于分类分数排名前 n 的患者中。在多分类问题中,recall@1 的平均值等同于平衡准确率。此外,我们还提供了对每个数据集在评估 N 名患者时,随机选择正确患者的概率。
5.1 实验1的结果在三个数据集中,基于组织学切片的患者再识别方法均表现出令人满意的性能,详细结果见表1。在MEN数据集中,结合UNI特征提取器的TransMIL分类器实现了最高的 recall@1,达到了77.09%。对于将WSI(全切片图像)图像块投影到潜在空间的不同特征编码器的比较结果显示,UNI模型在所有比较的WSI分类器中表现最优。总体而言,所有基于组织病理学数据预训练的特征提取器性能均优于基于ImageNet的基线。
在两个公开数据集上,我们也得到了满意的结果。采用TransMIL方法结合UNI特征提取器,在LUAD数据集上实现了 recall@1 为77.19%,在LSCC数据集上则达到了80.01%。表2展示了使用染色增强后的实验1结果。与表1中未使用增强的结果相比,所有模型的性能均有所下降。对于基于图像块和naive-MIL的方法,这种下降尤为显著,recall@1 分别从74.92%降至65.25%,从74.98%降至61.13%。相比之下,对于WSI分类器,这种下降不太明显。其中,CLAM模型的 recall@1 降幅最大,从66.54%降至62.97%,而TransMIL方法则保持了较高的 recall@1 值,为76.14%。
5.2 实验2的结果
当模型使用MEN数据集中最早的切除样本进行训练,并在后续切除样本上进行测试时,与实验1相比性能显著下降(见表3)。在所有比较方法中,TransMIL方法表现最佳,recall@1 为15.10%,recall@5 为29.25%。尽管结果较实验1明显偏低,但仍显著高于随机猜测的概率。
5.3 实验1和实验2的事后分析结果
图6展示了实验1和实验2中测试样本的潜在空间嵌入与其对应潜在空间锚点之间的L2距离。在两个实验中,正确分类的样本与其对应锚点的距离明显小于错误分类的样本。此外,测试样本与锚点之间的距离在实验1中显著小于实验2。
Figure
图

Fig. 1. Overview of randomly selected patches from the three datasets used. In contrast to our in-house meningioma dataset (MEN), the lung adenocarcinoma (LUAD) and lungsquamous cell carcinoma (LSCC) datasets originating from TCIA exhibit a more pronounced visual variance. Each patch covers an area of about 0.012 square millimeters

Fig. 2. Scheme of the tissue preparation procedure used to prepare the slides in thein-house meningioma (MEN) dataset. A resection can be divided into one or morecontainers, each of which can be further divided into one or more blocks. However,
only one slide from each block is included in the data set
图2. 自有脑膜瘤(MEN)数据集中用于制备切片的组织处理流程示意图。一个切除样本可以被分成一个或多个容器,每个容器又可以进一步分成一个或多个蜡块。然而,数据集中仅包含每个蜡块对应的一个切片。

Fig. 3. Scheme of how the online stain augmentation was applied in the naive-MILmodel. During training, each of the images within one bag was augmented separately.
图3. 在naive-MIL模型中应用在线染色增强的示意图。在训练过程中,同一包内的每张图像都被单独进行增强处理。

Fig. 4. Given are versions of the same patch to which different intensities of stainaugmentation were applied. A stain augmentation based on the Macenkos stainnormalization method was used. The non augmented patch is given in the center ofthe grid.
图4. 展示了对同一图像块应用不同强度染色增强后的版本。染色增强基于Macenkos染色归一化方法,未增强的图像块位于网格的中心。

Fig. 5. Overview of the experimental setup of Experiments 1 and 2. Experiment 1 involved a tenfold Monte Carlo cross-validation. In Experiment 2, the slides from the earliestresection were used for training, while all images from later resections were used in a hold-out test dataset. To increase the statistical validity of the results of Experiment 2, tenmodels for each algorithm were trained on ten randomly selected training and validation splits drawn from the earliest resection of each patient.
图5. 实验1和实验2的实验设置概览。实验1采用十折蒙特卡洛交叉验证方法。实验2中,来自最早切除手术的切片用于训练,而所有来自后续切除手术的图像被用于保留测试数据集。为了增加实验2结果的统计有效性,每种算法分别训练了十个模型,这些模型基于从每位患者最早切除手术中随机选择的训练集和验证集划分而得。

Fig. 6. Distances between test samples and their respective latent space anchors. Subfigure (a) shows the distances for Experiment 1 and sub-figure (b) shows the distancesfor Experiment 2. In general, correctly classified samples are closer to their respectivelatent space anchors
图6. 测试样本与其对应潜在空间锚点之间的距离。子图(a)展示了实验1的距离,子图(b)展示了实验2的距离。总体而言,正确分类的样本更接近其对应的潜在空间锚点。
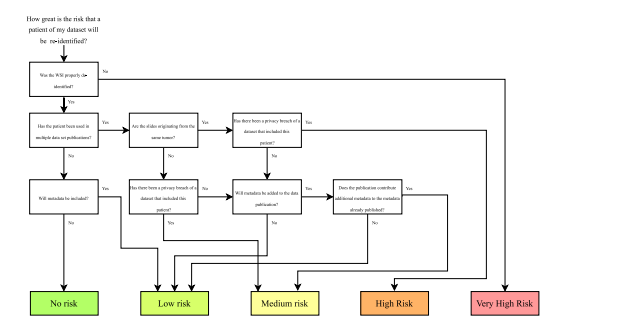
Fig. 7. Risk assessment scheme for estimating patient privacy risks when publishing histopathology images.
图7. 用于评估发布组织病理学图像时患者隐私风险的风险评估方案。
Table
表
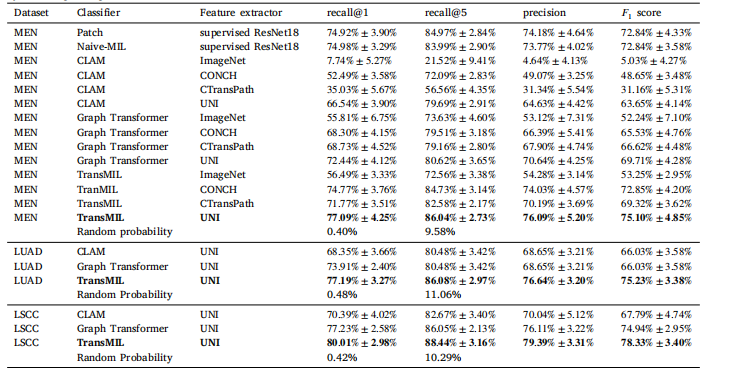
Table 1Results of Experiment 1. The respective means and standard deviations of the tenfold Monte Carlo cross-validation are given. In a multiclassclassification problem, the mean recall is equal to the balanced accuracy. Random probability is the probability of selecting the correct patientby random guessing
表1实验1的结果。表中给出了十折蒙特卡洛交叉验证的平均值和标准差。在多分类问题中,平均召回率等于平衡准确率。随机概率表示通过随机猜测选中正确患者的概率。

Table 2Results of Experiment 1 while using strong stain augmentation during training. The respective means and standard deviations of the tenfoldMonte Carlo cross-validation are given. In a multiclass classification problem, the mean recall is equal to the balanced accuracy. Randomprobability is the probability of selecting the correct patient by random guessing.
表2实验1在训练过程中使用强染色增强时的结果。表中给出了十折蒙特卡洛交叉验证的平均值和标准差。在多分类问题中,平均召回率等于平衡准确率。随机概率表示通过随机猜测选中正确患者的概率。

Table 3Results of Experiment 2. In a multiclass classification problem, the balanced accuracy equals the average recall. Random probability is theprobability of selecting the correct patient by random guessing
表3实验2的结果。在多分类问题中,平衡准确率等于平均召回率(recall)。

Table A.1Results of the preliminary investigation of the optimal magnification level for patch sampling. Given are the results of the tenfold Monte Carlocross-validation using the MEN dataset and the patch-based model. In each experiment, patches with a width and height of 512 pixels wereused. The spatial resolution is given in microns per pixel (mpp)
表A.1补充实验中关于图像块采样最佳放大倍数的结果。表中展示了基于MEN数据集和图像块模型的十折蒙特卡洛交叉验证结果。每次实验中,图像块的宽度和高度均为512像素。空间分辨率以每像素微米数(mpp)表示。