Title
题目
A Flow-based Truncated Denoising Diffusion Model for super-resolution Magnetic Resonance Spectroscopic Imaging
基于流的截断去噪扩散模型用于超分辨磁共振波谱成像
01
文献速递介绍
磁共振波谱成像(Magnetic Resonance Spectroscopic Imaging,MRSI)是一种研究体内代谢的非侵入性成像技术。由于代谢研究在理解包括神经退行性疾病、癌症和糖尿病等一系列疾病中具有关键作用,MRSI 已成为一种重要的临床和前沿研究工具。MRSI 可应用于任何具有核自旋的核素,而用于体内 MRSI 的最常见核素是质子(1H),因为其在代谢物中的核磁共振(NMR)灵敏度高且丰度高(De Graaf,2019)。
为了表征小病灶和病灶内的异质性,需要高空间分辨率的 MRSI。然而,由于代谢物浓度低所导致的时间和灵敏度限制,在实际操作中,MRSI 通常以低分辨率采集。在临床实践中,通常使用 3T 扫描仪,并要求扫描时间在可接受范围内。为了获得满意的信噪比(SNR),1H-MRSI 的空间分辨率通常被限制在大约 1×1×1 cm³。尽管硬件和加速技术的进步显著(Bogner et al., 2021),但通过更强磁场(例如 7T)或延长扫描时间实现高分辨率 MRSI 仍在很大程度上不适合常规临床应用,特别是在现有的 3T MRI 系统上。因此,开发一种后处理方法从低分辨率扫描生成高分辨率 MRSI 将极大地提升其临床应用价值。
传统的用于超分辨率 MRSI 的后处理方法主要采用基于模型的正则化方法,这些方法结合了来自对应 MRI 扫描的高分辨率解剖信息,并通过全变差距离(total variation distance)对空间平滑性进行约束(Lam 和 Liang, 2014;Jain 等, 2017;Kasten 等, 2016;Hangel 等, 2019)。然而,这些基于正则化的方法存在以下问题:(1) 虽然 MRI 强度和 MRSI 代谢物浓度之间存在相关性,但 MRI 缺乏代谢信息。因此,过度依赖基于 MRI 的正则化可能导致生成的高分辨率 MRSI 偏向 MRI(Dong 等, 2023)。(2) 手工设计的正则化方法(例如全变差距离)无法真实反映高分辨率 MRSI 的内在特性,可能导致结果过度平滑。(3) 每次新采集都需要进行优化过程,这非常耗时。
深度学习在医学成像超分辨率任务中展现出了巨大潜力(Li 等, 2021),并能有效规避传统技术的局限性。首个用于超分辨率 MRSI 的深度学习方法(Iqbal 等, 2019)利用 Dense UNet 将低分辨率 MRSI 代谢图映射为高分辨率图,同时结合了 T1 MRI 的解剖信息。然而,由于缺乏公共的 MRSI 训练数据集,这些代谢图是由 MRI 合成的,这存在一个根本问题,即合成的 MRSI 可能无法真实捕获所有代谢物特性。为了解决这一问题,Dong 等(2021)引入了一个从高级别胶质瘤患者中获取的体内 1H-MRSI 数据集,用于训练超分辨率 MRSI 网络。这项工作旨在通过应用对抗损失(adversarial loss)来恢复高分辨率细节并增强视觉质量,这也在后续研究中得到了探索(Dong 等, 2022b;Li 等, 2022b)。通过生成对抗网络(Generative Adversarial Networks,GAN)(Goodfellow 等, 2020)引入对抗损失(Wang 等, 2020;Chen 等, 2024a)通常会受到训练不稳定性和模式崩溃问题的影响。
为了更准确地学习高分辨率 MRSI 图像的分布,Dong 等(2022c)提出了一种基于正态化流(normalizing flow)的网络。该框架通过似然最大化(likelihood maximization)来学习目标分布,从而使训练更加稳定且可解释,并优于基于 GAN 的方法。然而,基于流的方法必须通过可逆神经网络(invertible neural networks)实现(Ardizzone 等, 2018;Lugmayr 等, 2020;Liang 等, 2021;Dong 等, 2022a;Chen 等, 2024b),这在架构上存在约束,并限制了学习能力。
Aastract
摘要
Magnetic Resonance Spectroscopic Imaging (MRSI) is a non-invasive imaging technique for studying metabolismand has become a crucial tool for understanding neurological diseases, cancers and diabetes. High spatialresolution MRSI is needed to characterize lesions, but in practice MRSI is acquired at low resolution due totime and sensitivity restrictions caused by the low metabolite concentrations. Therefore, there is an imperativeneed for a post-processing approach to generate high-resolution MRSI from low-resolution data that can beacquired fast and with high sensitivity. Deep learning-based super-resolution methods provided promisingresults for improving the spatial resolution of MRSI, but they still have limited capability to generate accurateand high-quality images. Recently, diffusion models have demonstrated superior learning capability than othergenerative models in various tasks, but sampling from diffusion models requires iterating through a largenumber of diffusion steps, which is time-consuming. This work introduces a Flow-based Truncated DenoisingDiffusion Model (FTDDM) for super-resolution MRSI, which shortens the diffusion process by truncating thediffusion chain, and the truncated steps are estimated using a normalizing flow-based network. The networkis conditioned on upscaling factors to enable multi-scale super-resolution. To train and evaluate the deeplearning models, we developed a1H-MRSI dataset acquired from 25 high-grade glioma patients. We demonstratethat FTDDM outperforms existing generative models while speeding up the sampling process by over 9-foldcompared to the baseline diffusion model. Neuroradiologists’ evaluations confirmed the clinical advantagesof our method, which also supports uncertainty estimation and sharpness adjustment, extending its potentialclinical applications
磁共振波谱成像(MRSI)是一种非侵入性成像技术,用于研究代谢活动,已成为理解神经疾病、癌症和糖尿病的重要工具。为了更好地表征病变,MRSI需要高空间分辨率,但由于代谢物浓度低,受时间和灵敏度限制,实际采集的MRSI通常是低分辨率的。因此,迫切需要一种后处理方法,从可以快速采集且灵敏度高的低分辨率数据生成高分辨率MRSI。基于深度学习的超分辨率方法在提高MRSI空间分辨率方面表现出了良好的效果,但在生成准确且高质量图像方面仍有一定局限性。近年来,扩散模型在多种任务中表现出比其他生成模型更强的学习能力,但从扩散模型中采样需要经历大量的扩散步骤,耗时较长。
本研究提出了一种基于流的截断去噪扩散模型(FTDDM),用于MRSI超分辨率。该模型通过截断扩散链来缩短扩散过程,截断步骤通过基于归一化流的网络进行估计。该网络以放大倍数为条件,实现多尺度超分辨率。为了训练和评估深度学习模型,我们开发了一个从25例高级别胶质瘤患者中采集的¹H-MRSI数据集。研究结果表明,与现有生成模型相比,FTDDM在提高性能的同时,将采样速度提高了9倍以上(与基线扩散模型相比)。神经放射科医生的评估确认了该方法的临床优势。此外,该方法还支持不确定性估计和清晰度调节,进一步拓展了其潜在的临床应用范围。
Method
方法
2.1. Dataset
3D high-resolution 1H-MRSI, T1-weighted (MP2RAGE) and FluidAttenuated Inversion Recovery (FLAIR) scans from 25 high-gradeglioma patients were acquired on a 7T whole body MRI (Magnetom,Siemens Healthcare) equipped with a 32-channel receive coil array(Nova Medical). IRB approval and informed consent from all participants were obtained. MRSI sequence used spatial–spectral encoding toacquire 3D metabolic maps with a 64 × 64 × 39 measurement matrixand 3.4 × 3.4 × 3.4 mm3 nominal resolution in 15 min (Hangel et al.,2020; Hingerl et al., 2020; Dong et al., 2021). This resolution is notablyhigh for MRSI due to the low SNR of metabolite signals. Additionalparameters included an acquisition delay of 1.3 ms, a repetition time(TR) of 450 ms and a spectral bandwidth of 2778 Hz.T1 and FLAIR MRIs were skull-stripped and co-registered to MRSIusing FSL v5.0 (Smith et al., 2004). The processed MRSI spectra, asdescribed in Hangel et al. (2020), Hingerl et al. (2020), were quantifiedusing LCModel v6.3-1 (Provencher, 2014) to obtain 3D metabolic maps.For each of these maps, voxels with insufficient quality (SNR < 2.5,or FWHM > 0.15 ppm due to B0 field inhomogeneity, or with severeextracranial lipid contamination proximal to the skull) were filteredout using a quality-filtering mask. In this work, we focus on 7 metabolites considered as major markers of onco-metabolism and proliferation (Hangel et al., 2020): total creatine (tCr), total choline (tCho), Nacetylaspartate (NAA), glutamate (Glu), inositol (Ins), glutamine (Gln),and glycine (Gly).From each 3D MRSI scan of the 25 patients, we selected 9∼18 axialslices with clear spatial characteristics of brain. Each slice provided7 metabolites, resulting in a total of 2275 2D metabolic maps. These64 × 64 metabolic maps are regarded as high-resolution ground truth,from which we derived low-resolution images using k-space truncation.The size of the truncation window depends on the upscaling factor,which will be discussed later.
2.1 数据集
3D高分辨率氢-1磁共振波谱成像(1H-MRSI)、T1加权(MP2RAGE)和液体衰减反转恢复(FLAIR)扫描数据来自25例高等级胶质瘤患者,这些数据通过7T全身磁共振成像仪(Magnetom,Siemens Healthcare)和32通道接收线圈阵列(Nova Medical)采集。研究获得了伦理委员会(IRB)的批准,并取得了所有参与者的知情同意。
MRSI序列采用空间-光谱编码,在15分钟内采集了分辨率为3.4 × 3.4 × 3.4 mm³,测量矩阵为64 × 64 × 39的3D代谢图像(参考Hangel et al., 2020; Hingerl et al., 2020; Dong et al., 2021)。这种分辨率在MRSI中较为罕见,因为代谢物信号的信噪比(SNR)较低。其他扫描参数包括1.3毫秒的采集延迟、450毫秒的重复时间(TR)和2778 Hz的频谱带宽。
T1和FLAIR磁共振图像通过FSL v5.0(Smith et al., 2004)软件进行颅骨去除并配准到MRSI。根据Hangel等(2020)和Hingerl等(2020)描述的处理方法,MRSI光谱使用LCModel v6.3-1(Provencher, 2014)进行定量分析,生成3D代谢图像。对于这些图像,质量不佳的体素(信噪比 < 2.5,或由于B0场不均导致的半峰宽(FWHM) > 0.15 ppm,或因靠近颅骨而受到严重脂质污染的体素)被质量筛选掩码过滤掉。
本研究关注7种被认为是肿瘤代谢和增殖重要标志物的代谢物(参考Hangel et al., 2020):总肌酸(tCr)、总胆碱(tCho)、乙酰天冬氨酸(NAA)、谷氨酸(Glu)、肌醇(Ins)、谷氨酰胺(Gln)和甘氨酸(Gly)。
从25名患者的每次3D MRSI扫描中,我们选择了9至18个具有明确脑部空间特征的轴位切片。每个切片提供7种代谢物数据,总计2275幅二维代谢图像。这些分辨率为64 × 64的代谢图像被视为高分辨率的真实值,通过k空间截断法衍生出低分辨率图像。截断窗口的大小取决于后续讨论的上采样因子。
Conclusion
结论
In this work, we introduce a novel Flow-based Truncated DenoisingDiffusion Model (FTDDM) for super-resolution MRSI. Our method truncates the diffusion chain and hence accelerates the sampling processof the diffusion model. The noisy image at the truncation point isestimated directly from Gaussian noise via a normalizing flow-basednetwork. The diffusion network is conditioned on the upscaling factor,facilitating multi-scale super-resolution. Additionally, a temperatureparameter is incorporated into our diffusion model to allow sharpnessadjustment. Experimental results on our self-developed in vivo 1HMRSI dataset indicate that FTDDM outperforms other deep learningmodels and achieves over 9-fold acceleration compared to the conventional diffusion model without loss of image quality. Neuroradiologists’assessments confirm the excellent image quality given by the proposedmethod from clinical perspectives and assist in identifying the lowestoperable resolution. To the best of our knowledge, we are the firstto develop deep learning-based super-resolution models and conductcomprehensive evaluations on in vivo MRSI dataset. Future work toimprove the generalization and robustness of the proposed methodcan involve the inclusion of more datasets acquired from other MRIsystems (Nassirpour et al., 2018; Dong et al., 2024) and/or frompatients with different tumor types. The proposed model can also beextended to deuterium metabolic imaging (DMI) (De Feyter et al., 2018;Dong et al., 2020).
在本研究中,我们提出了一种新颖的基于流的截断去噪扩散模型(FTDDM)用于超分辨率MRSI。我们的方法通过截断扩散链,从而加速了扩散模型的采样过程。截断点处的噪声图像是通过归一化流网络直接从高斯噪声估计得到的。扩散网络以放大因子为条件,促进了多尺度超分辨率。此外,我们的扩散模型中还引入了温度参数,以便调整图像的清晰度。通过在我们自行开发的体内1H MRSI数据集上的实验结果表明,FTDDM在不损失图像质量的情况下,比传统的扩散模型加速了超过9倍。神经放射科医师的评估确认了该方法从临床角度提供的优异图像质量,并帮助确定了可操作的最低分辨率。据我们所知,我们是首个基于深度学习的超分辨率模型,并在体内MRSI数据集上进行了全面评估。未来的工作可以通过加入更多来自其他MRI系统(Nassirpour等,2018;Dong等,2024)和/或不同肿瘤类型患者的数据集,来进一步提高该方法的泛化能力和鲁棒性。该模型也可以扩展到氘代代谢成像(DMI)(De Feyter等,2018;Dong等,2020)。
Results
结果
Table 1 presents the quantitative comparison of the proposed FTDDM against other deep learning models at three different upscalingfactors 𝑠 = 8.0, 4.0 and 2.0. Compared to the GAN-based model(MUNet-FS-cWGAN) and the flow-based model (Flow Enhancer), thediffusion model DDPM excels across all metrics. This highlights thesuperior learning capability of the diffusion model within this set ofgenerative models. However, DDPM requires 1000 NFE during sampling, translating to a sampling time of approximately 12.44 s for asingle image slice on our devices. Reducing the NFE to 100 with thesimple respacing method (𝑇𝑟𝑒𝑠𝑝𝑎𝑐𝑒 = 100) downgrades the performance,as skipping the diffusion steps yields images that are less denoised.Using the same number of sampling steps, DPM-Solver++ with 𝑇𝑠𝑜𝑙𝑣𝑒𝑟= 100 fails to perform better than the respacing method. However, weshow in Section 3.3.4 that DPM-Solver++ can outperform the respacingmethod when fewer sampling steps are used. The truncation methodTDPM at 𝑇𝑡𝑟𝑢𝑛𝑐 = 100 outperforms the respacing method and offers comparable performance with the DDPM at 𝑇 = 1000. TDPM requires 101NFE, due to an extra GAN function evaluation. Our proposed method,FTDDM, achieves even better performance than TDPM, indicating theflow-based network’s superiority over GAN in generating noisy imagesat the truncation point. For a single image slice, FTDDM at 𝑇**𝑡𝑟𝑢𝑛𝑐 =100 has a sampling time of approximately 1.33 s, which is over 9-fold acceleration relative to DDPM. Since evaluating the flow-basednetwork for generating noisy images at the truncation point takes extratime (one additional NFE), the speed increase is slightly below 10-fold. Wilcoxon signed-rank tests were conducted between each pair ofscores for all methods in Table 1, for which the results indicate that alldifferences are significant with P values < 0.001.
表 1 展示了在三种不同的放大因子 (𝑠 = 8.0, 4.0 和 2.0) 下,提出的 FTDDM 与其他深度学习模型的定量比较。与基于生成对抗网络 (GAN) 的模型 (MUNet-FS-cWGAN) 和基于流的方法 (Flow Enhancer) 相比,扩散模型 DDPM 在所有指标上表现出色。这表明扩散模型在该类生成模型中具有更强的学习能力。然而,DDPM 在采样过程中需要 1000 次函数评估 (NFE),这在我们的设备上意味着对单张图像切片的采样时间约为 12.44 秒。使用简单的重采样方法将 NFE 减少到 100 (𝑇𝑟𝑒𝑠𝑝𝑎𝑐𝑒 = 100) 会降低性能,因为跳过扩散步骤会导致生成的图像去噪效果较差。
在相同的采样步骤下,使用 𝑇𝑠𝑜𝑙𝑣𝑒𝑟 = 100 的 DPM-Solver++ 并未优于重采样方法。然而,如第 3.3.4 节所示,当使用更少的采样步骤时,DPM-Solver++ 可以优于重采样方法。截断方法 TDPM 在 𝑇**𝑡𝑟𝑢𝑛𝑐 = 100 时优于重采样方法,并且其性能与 𝑇 = 1000 的 DDPM 相当。TDPM 需要 101 次 NFE,因为包含了一次额外的 GAN 函数评估。我们提出的 FTDDM 方法表现甚至优于 TDPM,表明基于流的网络在生成截断点的噪声图像时优于 GAN。在 𝑇𝑡𝑟𝑢𝑛𝑐 = 100 时,FTDDM 对单张图像切片的采样时间约为 1.33 秒,相较于 DDPM 提升了 9 倍以上的加速效果。由于基于流的网络在生成截断点噪声图像时需要额外的时间(一次额外的 NFE),速度提升略低于 10 倍。
对表 1 中所有方法的评分进行了 Wilcoxon 符号秩检验。结果表明,所有方法之间的差异均显著,P 值 < 0.001。
Figure
图

Fig. 1. A comparison between (a) the conventional diffusion model DDPM and (b) ourmethod FTDDM. 𝐱0 is the noiseless high-resolution MRSI metabolic map. The forwarddiffusion process gradually adds Gaussian noise to 𝐱0 . The noise is only added tothe region of interest, as defined by a quality-filtering mask, to avoid the necessityof suppressing noise from the background during the reverse diffusion process. Thereverse diffusion process uses a denoising network with parameters 𝜃 to retrace theforward diffusion process, provided with any condition images 𝐲. 𝐹𝜙 −1 is the inverse ofa normalizing flow-based network used to bridge the gap between the pure Gaussiannoise 𝐳 and the noisy image at the truncation point 𝐱𝑇𝑡𝑟𝑢𝑛𝑐
图 1. 对比 (a) 常规扩散模型 DDPM 与 (b) 我们的方法 FTDDM。𝐱₀ 表示无噪声的高分辨率 MRSI 代谢图。正向扩散过程逐渐向 𝐱₀ 添加高斯噪声。噪声仅添加到感兴趣区域(由质量筛选掩模定义),从而在逆向扩散过程中无需抑制背景噪声。逆向扩散过程通过带有参数 𝜃 的去噪网络来回溯正向扩散过程,同时结合任意条件图像 𝐲。𝐹𝜙⁻¹ 表示基于正态化流的网络的逆运算,用于弥合纯高斯噪声 𝐳 与截断点 𝐱*𝑇𝑡𝑟𝑢𝑛𝑐 的噪声图像之间的差距。
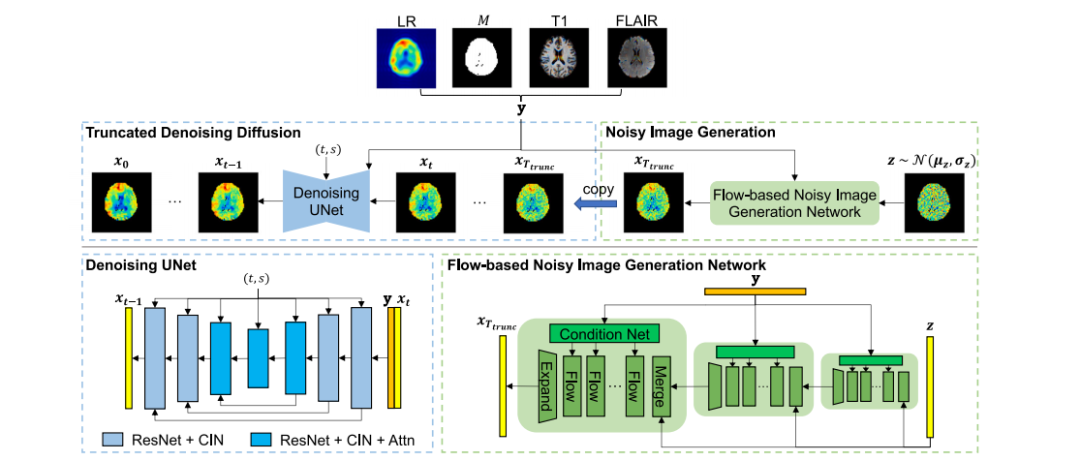
Fig. 2. Overview of the proposed method. The truncated denoising diffusion employs a Denoising UNet to iteratively estimate and remove noise from 𝐱𝑇𝑡𝑟𝑢𝑛𝑐, resulting in a noiselesshigh-resolution MRSI metabolic map 𝐱0 . The Denoising UNet also takes the condition 𝐲, which is a concatenation of the low-resolution (LR) metabolic map, a quality-filtering mask𝑀, T1 MRI and FLAIR MRI, i.e. 𝐲 = {LR, 𝑀, T1, FLAIR}. The Denoising UNet consists of Residual Network (ResNet) blocks and Conditional Instance Normalization (CIN). TheCIN embeds timestep 𝑡 and upscaling factor 𝑠 into the network. The blocks in the middle have multi-head attention (Attn) modules, following Nichol and Dhariwal (2021). 𝐱𝑇𝑡𝑟𝑢𝑛𝑐is generated from the Gaussian noise 𝐳 via the flow-based noisy image generation network, which comprises a series of flow layers across multiple dimensions, in line with Donget al. (2022c). Each flow layer contains conditional affine coupling, affine injector, invertible 1 × 1 convolution and activation normalization (Lugmayr et al., 2020). The conditionimages 𝐲 are infused into the flow layers through Condition Networks (Condition Net), which consist of convolution layers and LeakyReLU.
图 2. 所提方法的概述。截断去噪扩散模型通过一个去噪UNet迭代估计并去除 𝐱𝑇𝑡𝑟𝑢𝑛𝑐 中的噪声,生成无噪声的高分辨率MRSI代谢图 𝐱0 。去噪UNet还接收条件输入 𝐲,其由低分辨率(LR)代谢图、质量筛选掩码 𝑀、T1 MRI 和 FLAIR MRI 组成,即 𝐲 = {LR, 𝑀, T1, FLAIR}。去噪UNet包含残差网络(ResNet)块和条件实例归一化(CIN)。CIN 将时间步长 𝑡 和上采样因子 𝑠 嵌入网络。中间的模块包含多头注意力(Attn)模块,遵循 Nichol 和 Dhariwal(2021)的设计。𝐱𝑇𝑡𝑟𝑢𝑛𝑐 是通过基于流的噪声图像生成网络从高斯噪声 𝐳 生成的。该网络由多个维度上的一系列流层组成,与 Dong 等(2022c)一致。每个流层包含条件仿射耦合、仿射注入、可逆 1×1 卷积和激活归一化(参考 Lugmayr 等,2020)。条件图像 𝐲 通过条件网络(Condition Net)注入流层中,条件网络由卷积层和 LeakyReLU 构成。

Fig. 3. Qualitative comparisons of FTDDM against other methods at upscaling factor 𝑠 = 4.0. The two examples are: a tCr image from patient p1 and a Gly image from patient p2.FLAIR MRI provides the corresponding anatomical reference, with the tumor delineated by the red dashed line. Each metabolic map is shown alongside with its error map, exceptfor ground truth. Note that the images below ground truth, framed in red, are the standard deviation maps of 50 FTDDM samples and can be used for uncertainty estimation(they are not error maps of ground truth).
图 3. FTDDM 与其他方法在上采样因子 𝑠 = 4.0 下的定性比较。两个示例分别为:患者 p1 的 tCr 图像和患者 p2 的 Gly 图像。FLAIR MRI 提供了相应的解剖参考,肿瘤通过红色虚线标出。每个代谢图像旁边显示其误差图,除了地面真实值。请注意,地面真实值下方的图像框中显示的是50个 FTDDM 样本的标准差图,可以用于不确定性估计(它们不是地面真实值的误差图)。
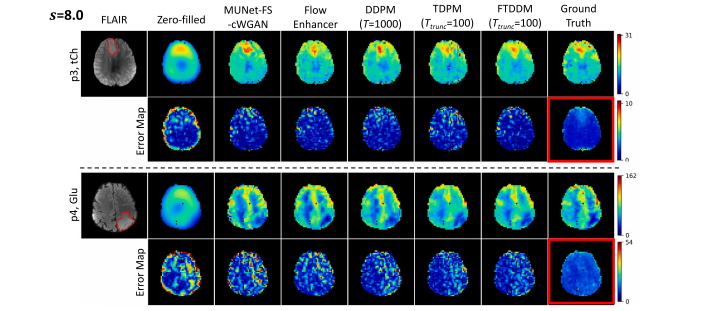
Fig. 4. Qualitative comparisons of FTDDM against other methods at upscaling factor 𝑠 = 8.0. The two examples are: a tCh image from patient p3 and a Glu image from patientp4.
图 4. FTDDM 与其他方法在上采样因子 𝑠 = 8.0 下的定性比较。两个示例分别为:患者 p3 的 tCh 图像和患者 p4 的 Glu 图像。

Fig. 5. Model performance of DDPM, DDPM with respacing, DPM-Solver++, TDPMand FTDDM at different numbers of sampling steps (𝑇𝑟𝑒𝑠𝑝𝑎𝑐𝑒 for DDPM respace, 𝑇𝑠𝑜𝑙𝑣𝑒𝑟for DPM-Solver++, 𝑇𝑡𝑟𝑢𝑛𝑐 for TDPM and FTDDM).
图 5. 不同采样步数下,DDPM、带重采样的DDPM、DPM-Solver++、TDPM和FTDDM模型的性能。(𝑇𝑟𝑒𝑠𝑝𝑎𝑐𝑒 表示DDPM重采样的步数,𝑇𝑠𝑜𝑙𝑣𝑒𝑟 表示DPM-Solver++的步数,𝑇𝑡𝑟𝑢𝑛𝑐 表示TDPM和FTDDM的步数)。

Fig. 6. Model performance (measured in SSIM) under different combinations of actualupscaling factors (horizontal axis) and conditioned upscaling factors (color bars). Forconciseness, only 5 adjacent values of conditions are shown for each actual upscalingfactor
图 6. 在不同实际上采样因子(横轴)和条件上采样因子(颜色条)组合下的模型性能(以SSIM为衡量标准)。为简洁起见,每个实际上采样因子仅显示了5个相邻的条件值。

Fig. 7. Sharpness adjustment with 𝜏𝑑 . (a) Network performance in PSNR, SSIM andLPIPS at various levels of 𝜏𝑑 . Lower LPIPS (↓) is better. (b) Visual sharpness of asuper-resolution Ins image at various 𝜏𝑑 compared to ground truth (GT). The tumor isdelineated by the red dashed line in FLAIR
图 7. 使用 𝜏𝑑 进行锐度调整。(a) 在不同 𝜏𝑑 水平下,网络在PSNR、SSIM和LPIPS上的表现。较低的LPIPS(↓)表示效果更好。(b) 在不同 𝜏𝑑 下,超分辨率的肌醇(Ins)图像与地面真实图像(GT)进行对比,FLAIR图像中肿瘤由红色虚线标出。

Fig. 8. Image quality assessment based on neuroradiologists’ ratings. Image quality of low-resolution (LR) images and super-resolution (SR) images given by FTDDM was blindreviewed by neuroradiologist 1 and 2. The horizontal axis shows the evaluation metrics. Each colored bar denotes the count of images that received a specific score for thecorresponding metric. The scoring system is: NA (not applicable), 10 (poor), 30 (bad), 50 (fair), 70 (good) and 90 (excellent).
图 8. 基于神经放射科医生评分的图像质量评估。低分辨率(LR)图像和由FTDDM生成的超分辨率(SR)图像经过神经放射科医生1和2的盲审。水平轴表示评估指标。每个彩色条形图表示在相应指标下获得特定分数的图像数量。评分系统为:NA(不适用)、10(差)、30(不好)、50(一般)、70(好)和90(优秀)。

Fig. 9. Visualization of low-resolution (zero-filled from 8 × 8), FTDDM super-resolutionand ground truth images of 7 metabolites from a slice containing tumor. The first rowshows T1 MRI, FLAIR MRI and an example of quality-filtering mask for NAA. Thetumor is delineated by the red dashed line in FLAIR
图 9. 低分辨率(从8 × 8零填充)、FTDDM超分辨率图像与真实图像的可视化,来自包含肿瘤的切片中的7种代谢物。第一行展示了T1 MRI、FLAIR MRI和NAA的质量过滤掩膜示例。肿瘤在FLAIR图像中由红色虚线标出。

Fig. 10. Image quality assessment for three extremely low resolutions 2 × 2, 4 × 4 and 6 × 6. Due to limited space, only a subset of most representative evaluation metrics isdisplayed. For each metric shown on the horizontal axis, ratings for low-resolution (LR) images at 2 × 2, 4 × 4 and 6 × 6 (indicated by the number in the parenthesis) and theircorresponding super-resolution (SR) images are presented.
图 10. 三种极低分辨率(2 × 2、4 × 4 和 6 × 6)图像质量评估。由于空间限制,仅显示了最具代表性的评估指标子集。每个横轴上的指标,展示了低分辨率(LR)图像在2 × 2、4 × 4和6 × 6(由括号中的数字表示)下的评分,以及它们对应的超分辨率(SR)图像的评分。
Table
表

Table 1Quantitative comparisons of FTDDM against other deep learning methods at three upscaling factors 𝑠 = 8.0, 4.0, 2.0. NFE: number of function evaluations, Time: Sampling time inseconds (s) for a single image slice, Params: number of model parameters in millions (M), NRMSE: Normalized Root Mean Square Error, PSNR: Peak Signal-to-Noise Ratio, SSIM:Structural Similarity Index.
表 1FTDDM 与其他深度学习方法在三个上采样因子 𝑠 = 8.0、4.0、2.0 下的定量比较。NFE:函数评估次数,时间:单张图像切片的采样时间(秒),参数:模型参数数量(百万),NRMSE:归一化均方根误差,PSNR:峰值信噪比,SSIM:结构相似性指数。

Table 2Ablation studies on the diffusion model and the normalizing flow model in FTDDM.
表 2FTDDM 中扩散模型和归一化流模型的消融研究。

Table 3Ablation studies on design elements. ✓ or × indicates whether a certain design elementis included or not.
表 3设计元素的消融研究。✓ 或 × 表示某个设计元素是否包含。

Table 4Study of combining DPM-Solver++ with FTDDM to further reduce the number of sampling steps. SSIM scores are shown foreach valid combination of 𝑇𝑡𝑟𝑢𝑛𝑐 in FTDDM and 𝑇**𝑠𝑜𝑙𝑣𝑒𝑟 in DPM-Solver++
表 4结合DPM-Solver++与FTDDM以进一步减少采样步数的研究。展示了FTDDM中𝑇𝑡𝑟𝑢𝑛𝑐和DPM-Solver++中𝑇𝑠𝑜𝑙𝑣𝑒𝑟的每种有效组合的SSIM评分。