Title
题目
Space-feature measures on meshes for mapping spatial transcriptomics
用于空间转录组学映射的网格空间特征度量
01
文献速递介绍
我们正经历一个以方法驱动的神经解剖学复兴期,其特点是大型项目的推动,产生了前所未有的、跨多个互补模态的高分辨率大脑空间数据。近年来,在小鼠大脑解剖结构和功能的成像方面,开发高度自动化的显微仪器取得了许多进展(Narasimhan等, 2017; Osten 和 Margrie, 2013; Ragan等, 2012; Zheng等, 2013),包括密集三维电子显微镜(EM)中的形态重建(Helmstaedter, 2013)以及全脑重建的中尺度(如 BRAIN 细胞普查网络 (BICCN) 项目 Ecker等, 2017)、神经元投射组(Oh等, 2014; Zingg等, 2014)和全脑细胞类型分布图(Kim等, 2017)。
此外,空间转录组学技术(如 MERFISH (Chen等, 2015; Moffitt等, 2018; Xia等, 2019) 和 STARmap (Kebschull等, 2020; Wang等, 2018))能够同时生成数千种基因的海量表达数据;新兴的条形码技术也在将神经元投射组数据与单细胞水平的密集转录分析联系起来(Chen等, 2019; Sun等, 2021; Kebschull等, 2016)。
Aastract
摘要
Advances in the development of largely automated microscopy methods such as MERFISH for imagingcellular structures in mouse brains are providing spatial detection of micron resolution gene expression.While there has been tremendous progress made in the field of Computational Anatomy (CA) to performdiffeomorphic mapping technologies at the tissue scales for advanced neuroinformatic studies in commoncoordinates, integration of molecular- and cellular-scale populations through statistical averaging via commoncoordinates remains yet unattained. This paper describes the first set of algorithms for calculating geodesicsin the space of diffeomorphisms, what we term space-feature-measure LDDMM, extending the family of largedeformation diffeomorphic metric mapping (LDDMM) algorithms to accommodate a space-feature action onmarked particles which extends consistently to the tissue scales. It leads to the derivation of a cross-modalityalignment algorithm of transcriptomic data to common coordinate systems attached to standard atlases.We represent the brain data as geometric measures, termed as space-feature measures supported by a largenumber of unstructured points, each point representing a small volume in space and carrying a list of densitiesof features elements of a high-dimensional feature space. The shape of space-feature measure brain spaces ismeasured by transforming them by diffeomorphisms. The metric between these measures is obtained afterembedding these objects in a linear space equipped with the norm, yielding a so-called ‘‘chordal metric’’.
开发高度自动化显微方法(如 MERFISH)以成像小鼠大脑中的细胞结构的进展,为微米级分辨率的基因表达空间检测提供了支持。尽管在计算解剖学(CA)领域取得了显著进展,能够在组织尺度上进行微分同胚映射技术以在常见坐标系中开展高级神经信息学研究,但通过常见坐标系对分子和细胞尺度群体进行统计平均整合仍未实现。本文描述了第一套用于在微分同胚空间中计算测地线的算法,我们称之为空间特征度量 LDDMM,扩展了大变形同胚度量映射(LDDMM)算法家族,以适应对标记粒子的空间特征作用,且可一致地扩展到组织尺度。该算法引导了转录组数据跨模态对齐至标准图谱所附的公共坐标系的推导。
我们将大脑数据表示为几何度量,称为空间特征度量,其由大量非结构化点支撑,每个点代表空间中的一个小体积,并携带高维特征空间中特征元素的密度列表。通过对这些度量进行微分同胚变换来测量大脑空间的空间特征度量形状。在将这些对象嵌入带有规范的线性空间后获得这些度量之间的距离,形成所谓的“弦度量”。
Method
方法
Let denote a ‘‘feature’’ space, where features typically correspondto high-dimensional measurements made by an imaging system, describing biological function. We are interested in the combined analysisof space and function, and will therefore work with the product spaceR𝑑 × (𝑑 = 2 or 3).
设 表示“特征”空间,其中特征通常对应由成像系统进行的高维测量,描述生物功能。我们关注空间和功能的联合分析,因此将使用乘积空间 R𝑑 × (𝑑 = 2 或 3)。
Figure
图
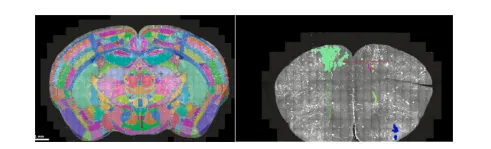
Fig. 1. Depicting Allen Atlas scale (left) and mouse micro scales right.
图 1. 展示了 Allen 图谱尺度(左)和小鼠微观尺度(右)。

Fig. 2. Mai–Paxinos atlas section and high field MRI with Tau molecular pathology (right).
图.2.马-帕克斯诺斯脑部切片图和高场强磁共振成像与tau分子病理学(右侧)。

Fig. 3. First row: First image component (values between 0 and 1) of the 2D template and three targets with low, medium and high noise. Second row: Deformed templates afterrunning LDDMM. Third row: logarithm of the Jacobian determinant of the deformation (red: expansion; blue: compression).
图.3. 第一行:2D模板中第一个图像组件(值在0到1之间)以及三个具有低、中、高噪声的目标。第二行:经过变形的模板运行LDDMM算法。第三行:变形的Jacobian行列式的对数(红色:膨胀;蓝色:压缩)。

Fig. 4. First row: First image component (values between 0 and 1) of the 3D template and two targets with low and high noise. Second row: Deformed templates after runningLDDMM. Third row: logarithm of the Jacobian determinant of the deformation (red: expansion; blue: compression).
图 4. 第一行:3D 模板的第一个图像分量(值在 0 到 1 之间)以及两个具有低噪声和高噪声的目标。第二行:运行 LDDMM 后的变形模板。第三行:形变的 Jacobian 行列式的对数(红色:扩张;蓝色:压缩)。

Fig. 5. MERFISH images (same mouse, nearby slices): Top row, from left to right: template with gene provided by Atp6ap1l gene counts, target of a second mouse with the samegene, template mapped based on a total of 10 genes of highest variance to the second mouse target. Second row provides the same information with Atp6ap1l replaced by Satb2.
图 5. MERFISH 图像(同一只小鼠,相邻切片):第一行,从左到右依次为:由 Atp6ap1l 基因计数提供的模板,同一基因的另一只小鼠的目标,基于方差最高的 10 个基因映射到第二只小鼠目标的模板。第二行提供相同信息,但将 Atp6ap1l 替换为 Satb2。

Fig. 6. MERFISH images (different mice, visually matched slices): Top row, from left to right: template with gene provided by Atp6ap1l gene counts, target of a second mousewith the same gene, template mapped based on a total of 10 genes of highest variance to the second mouse target. Second row provides the same information with Atp6ap1lreplaced by Satb2. The first column (template) is the same as in Fig. 5.
图 6. MERFISH 图像(不同小鼠,视觉匹配的切片):第一行,从左到右依次为:由 Atp6ap1l 基因计数提供的模板,同一基因的另一只小鼠的目标,基于方差最高的 10 个基因映射到第二只小鼠目标的模板。第二行提供相同信息,但将 Atp6ap1l 替换为 Satb2。第一列(模板)与图 5 中相同。

Fig. 7. Deformation grids for the two mappings shown above, Figures Fig. 5, Fig. 6.Source: Data from Zeng (2022)
图 7. 上述映射(图 5 和图 6)的形变网格。

Fig. 8. Comparison between measure matching and image matching. First row is measure matching with, from left to right: Template image, noisy target image, deformed template,log Jacobian determinant of the transformation. Second row is LDDMM velocity-based matching with, from left to right: blurred template and target images used as inputs ofregistration, deformed blurred template, deformation applied to the original unblurred template, log Jacobian determinant.
图 8. 度量匹配与图像匹配的对比。第一行是度量匹配,从左到右依次为:模板图像、带噪声的目标图像、变形后的模板、变换的 Jacobian 行列式对数。第二行是基于 LDDMM 速度的匹配,从左到右依次为:用作配准输入的模糊模板和目标图像、变形后的模糊模板、应用于原始非模糊模板的变形、Jacobian 行列式的对数。

Fig. 9. Atlas mapping: First row: Template (observed) and target (unobserved) labels. Second row: first and second feature probabilities for the deformed template (left, estimated) and the observed target (right, observed). Third row: same information for the third and fourth feature probabilities
图 9. 图谱映射:第一行:模板(观测)和目标(未观测)标签。第二行:变形模板的第一和第二特征概率(左,估计)以及观测到的目标(右,观测)。第三行:第三和第四特征概率的相同信息。